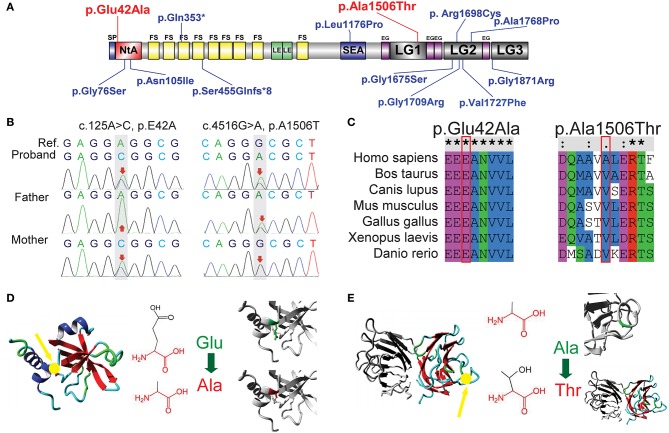
Figure 2.
Sequence analysis of two novel AGRN mutations. (A) Previously reported mutations in AGRN of CMS cases are highlighted in blue, whereas the two reported AGRN mutations of the present study are highlighted in red. Note that most genetic mutations are located in NtA, LG2, and LG3 domains. (B) Trio analysis (i.e., patient and patient's parents) of novel mutations in the present study. The proband c.125A>C (p.Glu42Ala) is from the asymptomatic mother with a heterozygous mutation, whereas the proband c.4516G>A (p.Ala1506Thr) is derived from the asymptomatic father with a heterozygous mutation. The reference (Ref) derives from a cDNA sequence from GenBank. (C) Sequence conservation analysis. c.125A>C (p.Glu42Ala) is located in an extremely conserved fragment, but c.4516G>A (p.Ala1506Thr) was not as strictly conserved as c.125A>C across species. (D) Protein structural-effects analysis of c.125A>C (p.Glu42Ala) via HOPE. The change to a neutrally charged alanine (highlighted in red) from a negatively charged residue of glutamic acid (highlighted in green) disturbs the formation of a salt bridge and abolishes its function. The location of p42 is indicted by a yellow arrow. (E) Protein structural-effects analysis of c.4516G>A (p.Ala1506Thr). A smaller and less hydrophobic mutant threonine (highlighted in red) residue might disturb the core structure of the LG1 domain. The location of p1506 is indicted by a yellow arrow. EG, EGF-like domain; FS, follistatin-like domain; LE, laminin EGF-like domain; LG1/ LG2/ LG3/, the first/second/third laminin G-like domain; NtA, N-terminal agrin domain; SEA, sea urchin sperm protein, enterokinase, and agrin domain.