Figure 2.
Reduced Endogenous YAP Target Gene Expression Accompanies the Induction of Pluripotency
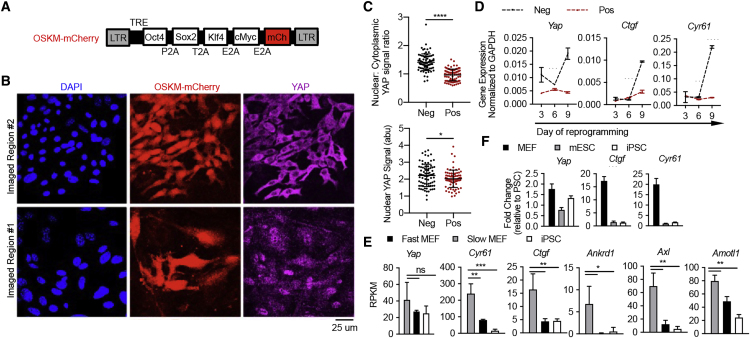
(A) Schema of polycistronic lentiviral vector encoding OSKM-mCherry.
(B) Primary MEFs transduced with OSKM-mCherry, treated with Dox for 4 days, and stained for YAP. Two representative regions in the same culture of distinct local cell density are shown.
(C) Quantification of endogenous YAP signal within mCherry– (Neg) and mCherry+ (Pos) cells, presented as a ratio of nuclear to cytoplasmic signal (top) or absolute nuclear signal (bottom; a.u., arbitrary unit). Nuclear area is defined as the DAPI+ region; cytoplasm is defined by pan-cellular GFP minus the DAPI+ region. Each dot denotes a single cell (n = 100 each).
(D) RT-qPCR analyses for endogenous Yap and YAP target genes Cyr61 and Ctgf in mCherry– (Neg) and mCherry+ (Pos) cells sorted after 3, 6, or 9 days of Dox treatment.
(E) Expression of endogenous Yap and YAP target genes Cyr61, Ctgf, Ankrd1, Axl, and Amot1 by mRNA sequencing in fast-cycling cells that exhibit enhanced reprogramming efficiency compared with slow-cycling cells and mature iPSCs. Data are re-plotted from (Guo et al., 2014) and are from three biological replicates. RPKM, reads per kilobase of transcript per million mapped reads.
(F) qRT-PCR of endogenous Yap, Ctgf, and Cyr61 in primary MEFs, mESCs, and iPSCs, displayed as the fold change relative to the average level in PSCs normalized to Gapdh. Data from three technical replicates are displayed in (D–F), representative of three independent experiments.