Figure 4.
MTLN Interacts with the Mitochondrial Energetics Machinery
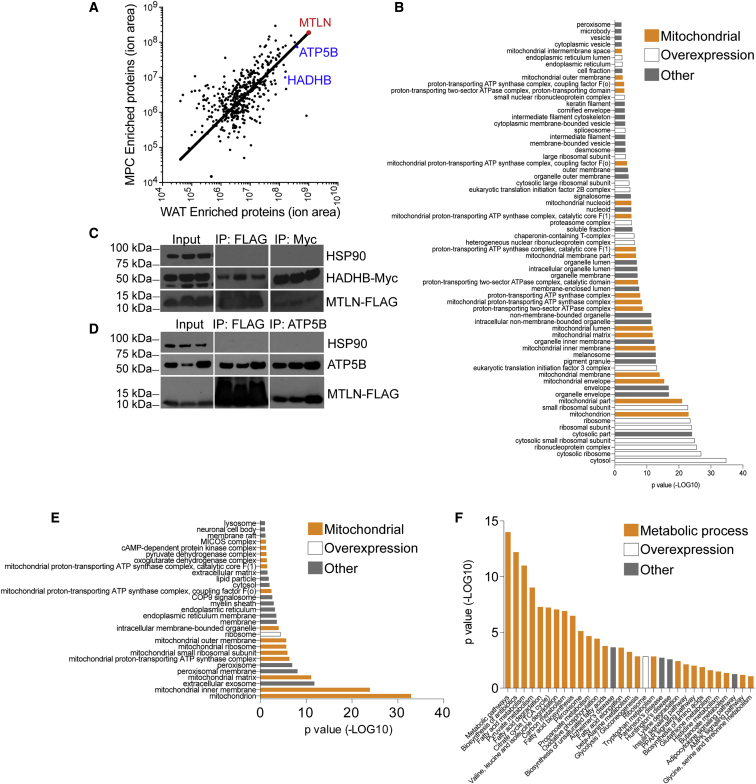
(A) Mass spectrometric profiling of co-immunoprecipitated proteins bound to MTLN-FLAG in undifferentiated MPCs and hPSC-adipocytes. Red, MTLN; blue, ATP5B and HADHB. Trendline represents the linear regression of these data using a non-linear model. Data based on two independent biological replicates each of MPCs and WAT. Data in Supplemental Table 2.
(B) DAVID gene ontology analysis of the cellular compartment categorization of co-immunoprecipitated proteins bound to MTLN-Flag. Orange, mitochondrial localization; white, protein translation machinery; gray, other. Modified Fisher exact p value, EASE score. Data in Supplemental Table 3.
(C) Immunoblot of HADHB co-immunoprecipitated with MTLN-FLAG. Whole-cell lysate is shown as input material.
(D) Immunoblot of ATP5B co-immunoprecipitated with MTLN-FLAG. Whole-cell lysate is shown as input material.
(E) DAVID gene ontology analysis of the cellular compartment categorization of co-immunoprecipitated proteins bound to MTLN-Flag in hPSC-adipocytes but not MPCs. Orange, mitochondrial localization; white, protein translation machinery; gray, other. Modified Fisher exact p value, EASE score.
(F) Gene ontology analysis describing the biological pathways enriched in the co-immunoprecipitated proteins bound to MTLN-Flag in hPSC-adipocytes but not in MPCs. Orange, mitochondrial processes; white, protein translation and overexpression; gray, other. Pathways and processes are colored through personal interpretation of DAVID terms. Modified Fisher exact p value, EASE score.
Related to Figure S4.