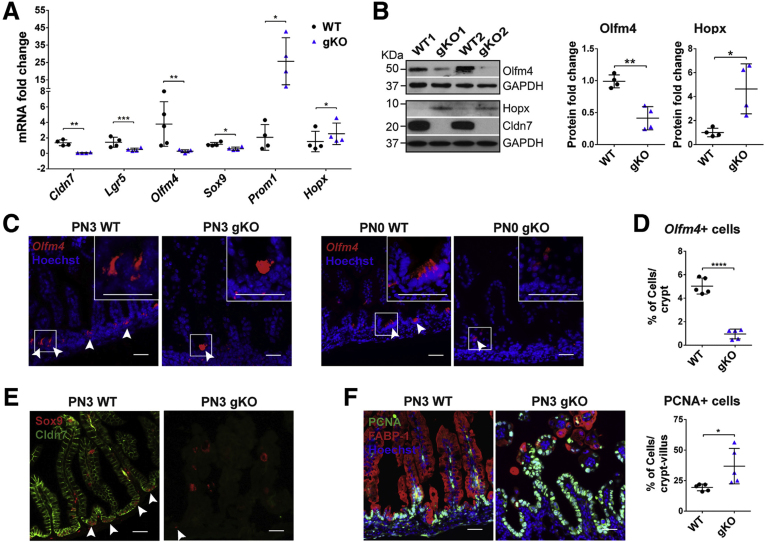
Figure 3.
Deletion of Cldn7 resulted in the loss of active crypt stem cells and an increase in epithelial cell proliferation in gKO SIs. (A) The qRT-PCR analysis of IESC marker genes in PN3 WT and gKO SIs. Lgr5, Olfm4, and Sox9 are active intestinal stem cell marker genes. Hopx is a quiescent stem cell marker gene and Prom1 is a transit-amplifying cell marker gene. n = 4–5; 2-tailed, paired t test; *P < .05, **P < .01, ***P < .001. (B) The protein levels of Olfm4 and Hopx were detected by immunoblotting (left) and quantified (right). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as a loading control. n = 4; 2-tailed, unpaired t test; *P < .05, **P < .01. (C) The IESC maker Olfm4 gene was detected by FISH in PN3 (left) and PN0 (right) WT and gKO SIs. Arrowheads indicate the Olfm4+cells. (D) Quantification of Olfm4+cells in panel C (PN3). n = 5; 2-tailed, unpaired t test; ****P < .0001. (E) Double-immunofluorescent labeling of Sox9 (red) and claudin-7 (green). Arrowheads indicate the Sox9+ cells. (F) Co-immunolabeling of proliferating cell marker PCNA (green) with enterocyte marker FABP-1 (red) (left). The quantification of PCNA-positive cells is shown (right). n = 5; 2-tailed, unpaired t test; *P < .05. Each point represents the calculation from 50 crypt–villus in each mouse. Data are represented as means ± SEM. Images are representative of more than 3 independent experiments. (C, E, and F) Scale bars: 100 μm.