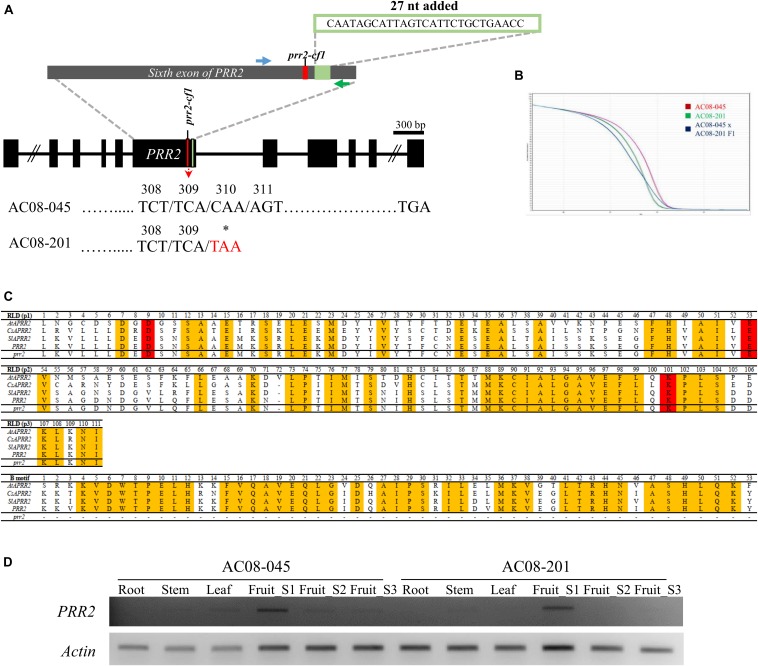
FIGURE 3.
Mutation and HRM markers of PRR2 in AC08-045 and AC08-201, alignment of PRR2 domain between Arabidopsis and three horticultural crops, and expression analysis of PRR2. (A) Comparison of PRR2 variation between AC08-045 and AC08-201. Variation in the nucleotide sequence and resulting codon change are highlighted in red. A premature stop codon due to a 1-bp substitution was detected in AC08-201. The sixth exon of PRR2 is expanded in the gray exon. Nucleotide sequence of 27-nt insertion observed in this study compared to Zunla v2.0 (Capana01g000809) is shown above. The positions of the designed HRM marker are shown in blue (forward primer) and green (reverse primer). Asterisks indicate stop codons. (B) HRM genotyping of PRR2. Melting curves are differentiated according to the accessions. Red, green, and blue lines indicate maternal, paternal, and heterozygous type of PRR2 genotype. x axis shows temperature and y axis shows normalized fluorescence. (C) Alignment of PRR2 domain between Arabidopsis and three horticultural crops. AtAPRR2, CsAPRR2, SlAPRR2, and PRR2 refer to the PRR2 genes of Arabidopsis thaliana, Cucumis sativus, Solanum lycopersicum, and Capsicum frutescens, respectively. RLD for receiver-like domain; B-motif for DNA-binding domain. (D) Expression pattern of PRR2 in various tissues. Results of RT-PCR amplification of PRR2. Roots, stems, leaves, and three stages of fruits were analyzed. In both AC08-045 and AC08-201, PRR2 was exclusively expressed in immature stage (Fruit_S1). Actin served as a control. S stands for stage.