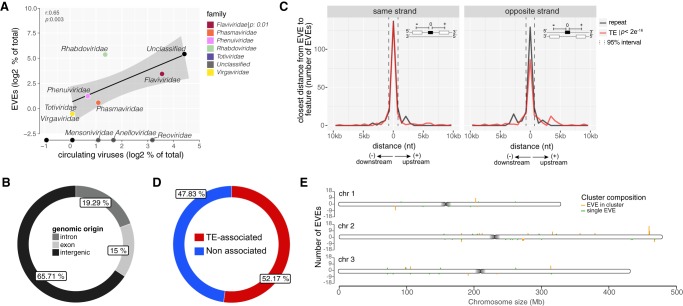
FIGURE 1.
Characterization of nonretroviral EVEs in A. aegypti mosquitoes. (A) Scatter plot showing the correlation between the abundance of viral families assigned to EVEs and exogenous viruses circulating in A. aegypti mosquitoes. r (Pearson correlation) and P-values are indicated. Viral families represented among EVEs are highlighted with different colors. Only the most abundant viral families exclusively found among circulating viruses are also named. (B) Genomic origin of EVEs in A. aegypti. (C) Distance of EVEs to the closest feature annotated in the same or opposite strands of the A. aegypti genome. Significance of colocalization was computed using Fisher's exact test for genomic data. A bin of 500 nt was utilized. EVEs annotated as genes were not considered for this analysis. (D) Percentage of EVEs association with TEs in the A. aegypti genome. EVEs found within 500 nt of TEs were considered associated. (E) Number of EVEs per chromosome of the AaegL5 version of the A. aegypti genome. EVEs were grouped by bins of 20 kb separated by strand. The AaegL5 version of the A. aegypti genome was used for EVE identification.