FIGURE 4.
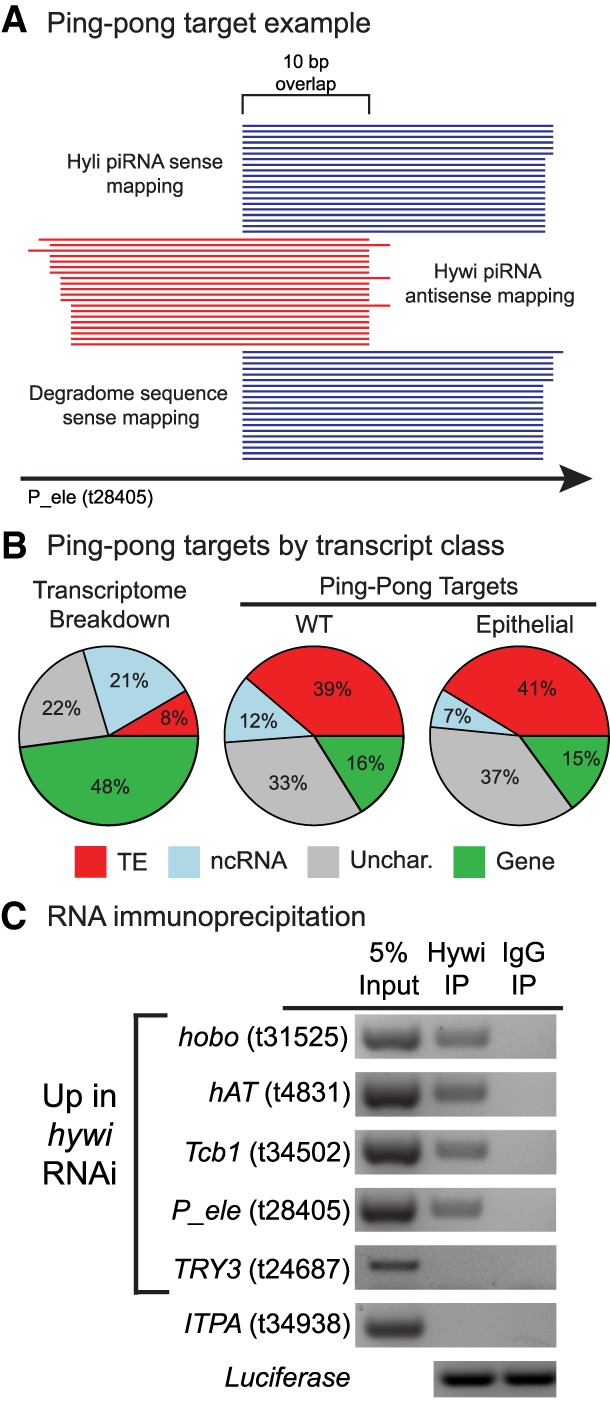
TE transcripts are cleaved by PIWI–piRNA complexes in Hydra ESCs. (A) Hywi-bound piRNAs, Hyli-bound piRNAs, and degradome reads were aligned to the transcriptome to identify likely direct targets of the PIWI–piRNA pathway. One example from the data is shown (t28405, P-element). The following pattern indicates cleavage: Antisense-oriented Hywi-bound piRNAs align with a 10-nt 5′ overlap with both sense-oriented Hyli-bound piRNAs and sense-oriented degradome reads. For a transcript to be considered a target of the PIWI–piRNA pathway, we require a minimum of 10 reads of each species to map in such an arrangement. (B) The degradome sequencing from WT Hydra identified 2047 transcripts (Supplemental Table S4) as targets of the PIWI–piRNA pathway, and the transcript class distribution of these targets is shown. The degradome sequencing from epithelial Hydra identified 254 transcripts (Supplemental Table S4) as targets of the PIWI–piRNA pathway, and the class distribution of these targets is shown. In contrast to the class distribution of the transcriptome as a whole, TE transcripts and uncharacterized transcripts comprise the majority of targets in both WT and epithelial animals. (C) RNA immunoprecipitation (RIP) was used to identify transcripts that directly interact with Hywi-piRNA complexes in ESCs (for complete results see Supplemental Fig. S7). Following RIP, luciferase mRNA was added to the washed beads to act as a positive control for RNA extraction and cDNA synthesis. RT-PCR was used to test for the presence of target transcripts in complexes immunoprecipitated with a Hywi antibody. The results for four TE transcripts are shown, which were amplified in three biological replicates (Supplemental Fig. S7). The following transcripts did not associate with Hywi protein as expected: (i) The TRY3 transcript (t24687) is up-regulated in response to hywi RNAi but has a low piRNA mapping density; and (ii) the ITPA transcript (t34938) did not change in response to hywi RNAi and has a low piRNA mapping density.
