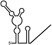
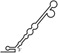
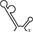
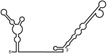
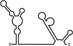
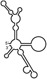
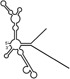
Table 1.
Summary of RdRp affinity and polymerase activity assays with various vRNA fragments
| RNAs | Poly(N) | SLA | SLA tail | SL | cySL | SLA–SL | SLA–cySL | cyUTRs | 5′ + 3′‐UTRs |
|---|---|---|---|---|---|---|---|---|---|
| 2D structures |

|
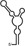
|
|
|
|
|
|
|
|
| Polymerase activity | Yes44 | No39 | Yes39 | No20, 21 | n.d. | No39 | Yes39 | Yes20, 21 | Yes (both 5′ and 3′ transcripts were observed)20, 21 |
| RdRp‐binding activity | No39 | Yes43 No39 | Yes39 | Yes40 No21 | Yes20, 40 | n.d. | n.d. | Yes40 | n.d. |
| Free 3′‐tail following the long stem | Yes | No | Yes | No | Yes | No | Yes | Yes | Yes |
n.d., not determined.
“cy” refers to structures as they would exist in cyclized form.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
