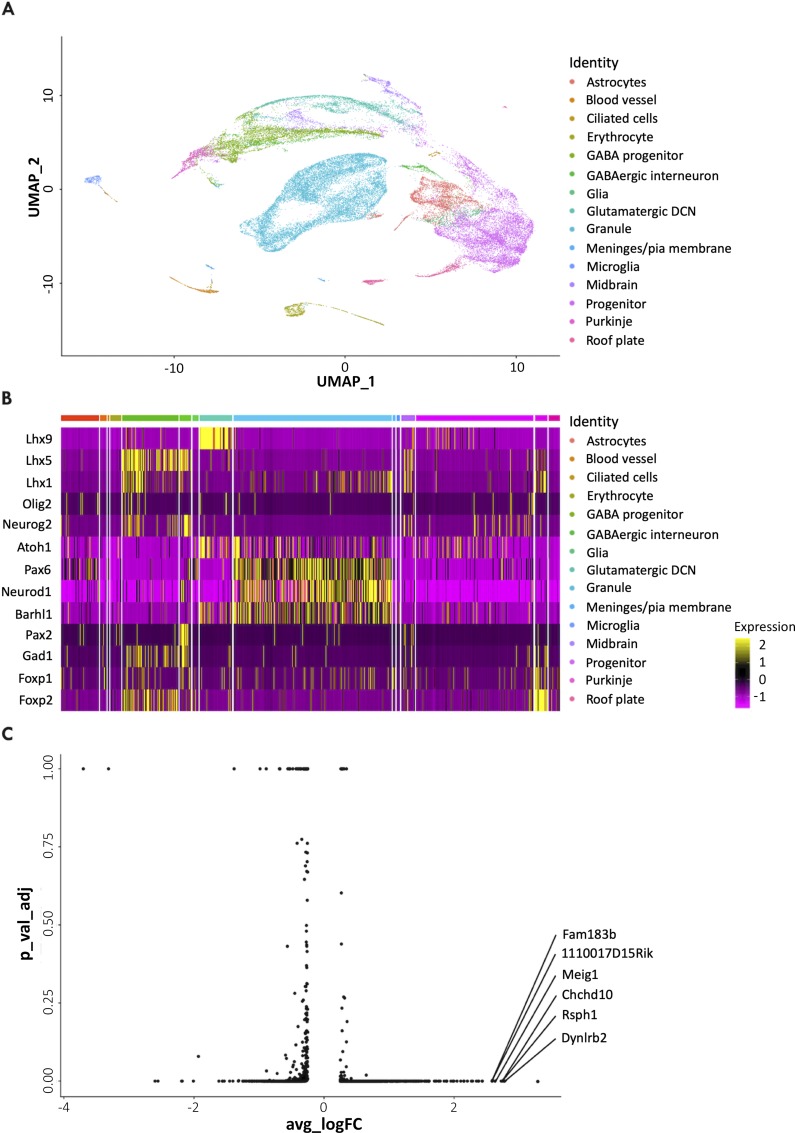
Fig 2. Single-cell characterisation of cellular subtypes in the mouse cerebellum.
Data from Carter and colleagues [36] was downloaded from the European Nucleotide Archive (PRJEB23051). In total, 39,245 cells meeting the authors’ quality control cutoffs were projected in UMAP space using Seurat v 3.1.0 according to the embedded metadata. The FindMarkers command was used to perform differential expression testing (ranked Wilcoxon sum test) to distinguish cell type–specific markers. Scaled data were visualised using the DoHeatmap command. (A) UMAP projection of the single-cell data set identifies all known major subtypes of the developing cerebellum. Nonneural cell types are also visualised. Differential gene expression at cellular resolution readily distinguishes the major cellular subtypes of the cerebellum (B) and identifies a novel cell type (C). (B) Heatmap of selected, established transcriptional markers of the respective cell types. Each vertical bar represents a single cell. Column (cell identity) width is proportional to the number of cells present in that cluster. (C) Volcano plot of differential gene expression in a rare cell type labelled ‘ciliated cells’. The labels indicate genes that encode ciliary proteins, which are significantly enriched in this cell type. Atoh1, atonal bHLH transcription factor 1; Barhl1, BarH like homeobox 1; Chchd10, coiled-coil-helix-coiled-coil-helix domain containing 10; DCN, deep cerebellar nuclear neuron; Dynlrb2, dynein light chain roadblock-type 2; Fam183b, family with sequence similarity 183, member B; Foxp, forkhead box P; Gad1, glutamate decarboxylase 1; Lhx, LIM homeobox protein; Meig1, meiosis expressed gene 1; Neurod1, neurogenic differentiation 1; Neurog2, neurogenin 2; Olig2, oligodendrocyte transcription factor 2; Pax, paired box; Rsph1, radial spoke head 1 homolog; UMAP, Uniform Manifold Approximation and Projection.