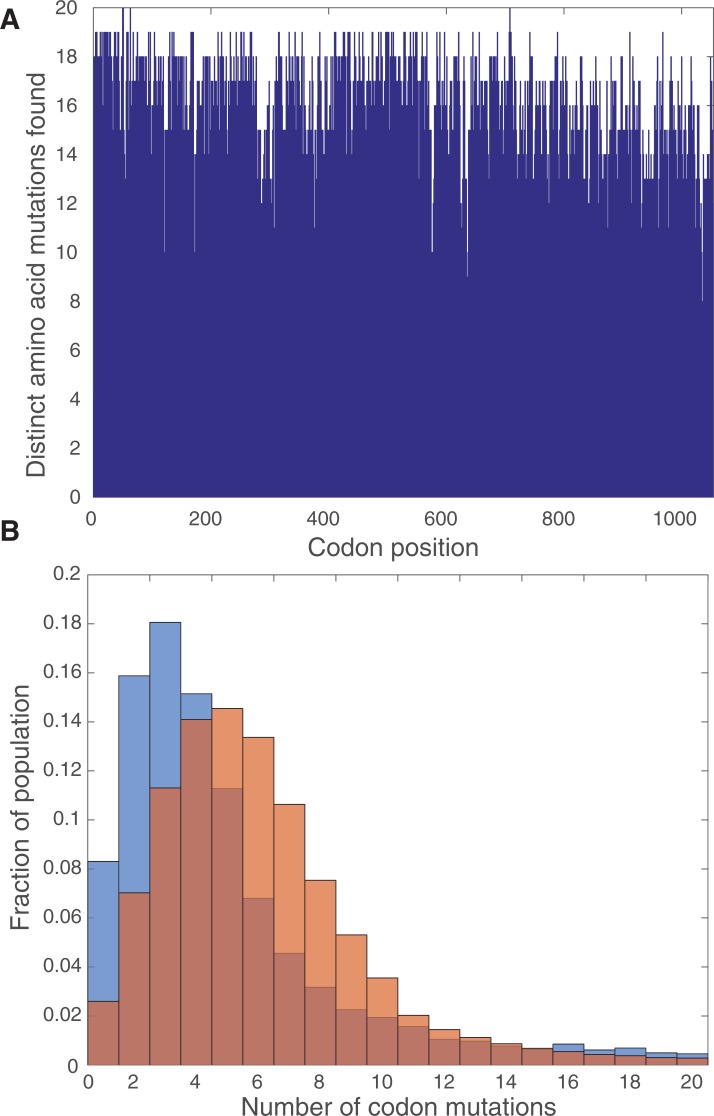
Fig 1. SMOOT can mutate all codons in a gene at a tunable mutation rate.
(A) SMOOT optimization was originally performed on SaCas9 before being applied to SpCas9. Distribution of distinct nonsynonymous mutations produced by SMOOT throughout the Cas9 sequence, as identified by PacBio long-read sequencing and pairwise sequence alignment and custom analysis via Matlab. Each distribution represents data from a single library. (B) Probability density function of number of codon mutations per mutant allele. The orange distribution was generated from a mutant library with no forward tuning primer added, while the blue distribution was generated from a library with 0.5 nM forward tuning primer added. Overlap of distributions is shown by intermediate color.