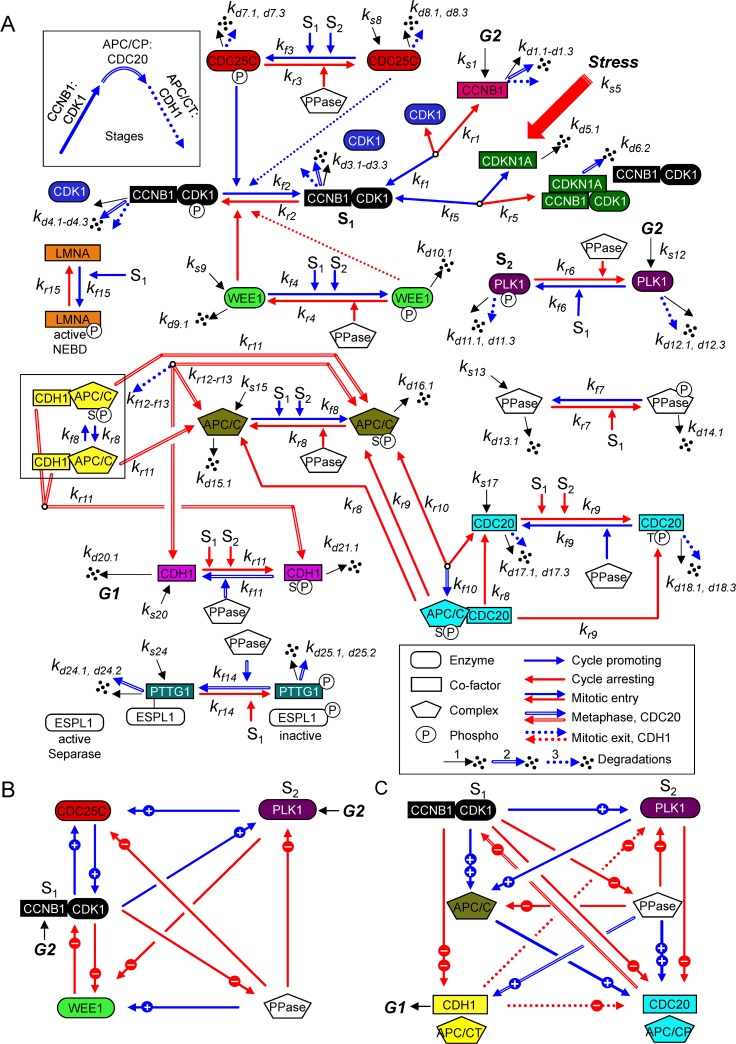
Fig 1. Regulation network of the mitotic cell cycle in human cells.
The mitotic biopathway for the reaction network of key mitotic proteins is converted into a mathematical structure based on ODEs involving the law of mass conservations and a hybrid framework combining mass action and Michaelis-Menten kinetics. (A) We have divided the biopathway into three stages (see inset) defined by arrow styles. Each factor, as defined in the text and Supplemental Information, is identified by a unique color conserved throughout the article. Major stages include: (1) Mitotic entry (solid arrows) involving CCNB1, CDK1, preMPF (CCNB1:CDK1P), MPF (CCNB1:CDK1), PLK1, WEE1, CDC25C, PPase (phosphatase), and LMNA, which is a substrate of MPF kinase (P, phosphorylated; NEBD, nuclear envelop breakdown); (2) Metaphase to anaphase transition (double arrows) involving APC/C:CDC20, PPase, and PTTG1 (Securin) in complex with ESPL1 (Separase); and (3) Mitotic exit (dashed arrows) involving APC/CT:CDH1 (T, total). Here, S1 denotes active MPF kinase (CCNB1:CDK1) and S2 denotes active PLK1 kinase (phosphorylated), which are major signals for many mitotic cell cycle reactions, as indicated. Reactions involving Cyclin-dependent kinase inhibitor CDKN1A (p21CIP1) have been included, and locations for G2 signals and exit into G1 are noted. The model consists of 26 ODEs governing 15 interaction reactions (kfn forward rate constant and krn reverse rate constant for reaction n) of 12 key mitotic proteins and associated protein complexes and includes synthesis (ks: synthesis rate constant) and multiple degradations (kdn.1: self-degradation rate constant, kdn.2: degradation rate constant by APCP/CDC20, and kdn.3: degradation rate constant by APC/CT:CDH1). (B, C) Simplified wiring diagrams based on (A), which identify both positive (blue, plus symbol) and negative (red, minus symbol) signals regulating the MPF kinase (CCNB1:CDK1) (B), and APC/CP:CDC20 and APC/CT:CDH1 ubiquitin ligase complexes with several signal strengths (C).