Abstract
Background
Deposition of complement factors on Mycobacterium leprae may enhance phagocytosis. Such deposition may occur through the lectin pathway of complement. Three proteins of the lectin pathway are produced from the gene MASP1: Mannan-binding lectin-associated serine protease 1 (MASP-1) and MASP-3 and mannan-binding lectin-associated protein of 44 kDa (MAp44). Despite their obvious importance, the roles played by these proteins have never been investigated in leprosy disease.
Methodology
We haplotyped five MASP1 polymorphisms by multiplex sequence-specific PCR (intronic rs7609662*G>A and rs13064994*C>T, exon 12 3’-untranslated rs72549262*C>G, rs1109452*C>T and rs850314*G>A) and measured MASP-1, MASP-3 and MAp44 serum levels in 196 leprosy patients (60%, lepromatous) and 193 controls.
Principal findings
Lower MASP-3 and MAp44 levels were observed in patients, compared with controls (P = 0.0002 and P<0.0001, respectively) and in lepromatous, compared with non-lepromatous patients (P = 0.008 and P = 0.002, respectively). Higher MASP-3 levels were present in controls carrying variants/haplotypes associated with leprosy resistance (rs13064994*T, rs1109452_rs850314*CG within GT_CCG and rs850314*A: OR = 0.5–0.6, Pcorr = 0.01–0.04). Controls with rs1109452*T, included in susceptibility haplotypes (GT_GTG/GT_CTG: OR = 2.0, Pcorr = 0.03), had higher MASP-1 and lower MASP-3 levels (P≤0.009). Those with GC_CCG, presented increasing susceptibility (OR = 1.7, Pcorr = 0.006) and higher MAp44 levels (P = 0.015). MASP-3 expression decreased in patients, compared with controls carrying rs1109452_rs850314*CA or CG (P≤0.02), which may rely on exon 12 CpG methylation and/or miR-2861/miR-3181 mRNA binding.
Conclusion
Polymorphisms regulating MASP-3/MAp44 availability in serum modulate leprosy susceptibility, underlining the importance of lectin pathway regulation against pathogens that exploit phagocytosis to parasitize host macrophages.
Author summary
Since immemorial times, Mycobacterium leprae inflicts permanent injuries in human kind, within a wide symptomatic spectrum ranging from insensitive skin patches to disabling physical lesions. Innate resistance to this parasite is well recognized, but poorly understood. The complement system is one of the most important arms of the innate response, and several lines of evidence indicate that it may be usurped by the parasite to enhance its entrance into host cells. These include our recent work on genetic association of the disease with lectin pathway components and the complement receptor CR1, whose polymorphisms modulate susceptibility to infection and clinical presentation. Here, we add another pivotal piece in the leprosy parasite-host interaction puzzle: polymorphisms and serum levels of three different lectin pathway proteins, all encoded by the same gene, namely mannan-binding lectin-associated serine protease 1 (MASP1). We found lower levels of two of these proteins, MASP-3 and MAp44, in leprosy patients. Higher MASP-3/lower MASP-1 levels were associated with protective haplotypes, containing two side-by-side polymorphisms located in the exclusive untranslated region of MASP-3 exon 12, which may regulate exon splicing and/or translation efficiency. The associations revealed in this study reflect the pleiotropic nature of this gene. They further illustrate the complexity of the response mounted against the parasite, which places MASP1 products in the regulatory crossroad between the innate and adaptive arms of the immunological system, modulating leprosy susceptibility.
Introduction
Leprosy is a chronic infectious disease caused by the obligate intracellular bacteria Mycobacterium leprae, with a 2018 global prevalence rate of 0.2/10,000 population, irreversibly disabling about 5.5% of 210,000 new cases every year. Brazil contributed to 92.3% of the increase in new American cases from 2017–18 [1]. Clinical manifestations depend on the host’s genetic polymorphisms and environmental factors that modulate the quality of the immune response, ranging from the multibacillary disabling lepromatous from one end of the clinical spectrum to the paucibacillary tuberculoid form at the other end, with borderline forms in between [2]. The very ancient origin of the complement system [3], made it an ideal platform to coevolve with pathogens, like M. leprae. To assist infection, M. leprae bacteria usurp complement activation to be opsonized and more readily phagocytosed into macrophages, one of their preferred host cells. In fact, the lectin pathway of complement (LP) has long been recognized as favoring the establishment of infection and development of disease [4–10]. There is also strong evidence that intracellular C3 cleavage directs T cell activation towards the Th1 pole, which is associated with the paucibacillary presentation of the disease. Since the first suggestion of balancing selection operating on the polymorphism of the gene encoding mannose-binding lectin (MBL2) due to protection against leprosy [4], much has been done investigating the possible roles played by LP genes and their products on the susceptibility to this disease [5–9] [11–15].
The lectin pathway of complement starts with the recognition of pathogen- or damaged/altered cell-associated patterns of carbohydrates or patterns of acetylated groups by pattern recognition molecules (PRMs i.e. collectins, MBL and ficolins—FCNs). These PRMs form circulating complexes with homodimers of MBL-associated serine proteases (MASPs) or MBL-associated proteins (MAps). Upon collectin/ficolin binding to a target, MASP-1 autoactivates and transactivates MASP-2, leading to the cleavage of complement factors C2 and C4, in order to form the C3 convertase (C4bC2b), also produced by the classical pathway. C4bC2b cleaves C3 and generates the anaphylatoxin C3a and the opsonin C3b. C3b is deposited on the target and recognized by complement receptors (CRs), which internalize the opsonized elements into phagocytes. Alternatively, formation of the C5 convertase (C4bC2bC3b) generates membrane pores (membrane attack complexes) formed by C5b and C6, C7, C8 and C9. The C3a and C5a anaphylatoxins further attract immune cells to the site of activation [16,17].
The MASP1 gene (3q27.3) encodes the serine proteases MASP-1 and MASP-3 and the non-enzymatic protein MAp44 (or MAP-1) [18]. These three proteins circulate in plasma as homodimers complexed with PRMs. The substrate specificity of MASP-1 is quite broad, resembling thrombin and trypsin. Besides cleaving MASP-2 in the lectin pathway, MASP-1 has procoagulant activity, cleaving and activating Factor XIII and fibrinopeptide, generating fibrinopeptide B and attracting neutrophils to assist the coagulation cascade [19]. It also activates carboxypeptidase B2, a molecule that prevents fibrinolysis and inactivates C3a and C5a anaphylatoxins [20]. MASP-1 generates bradykinin from the cleavage of high-molecular-weight kininogen [21]. It also cleaves PAR4 (protease-activated receptor 4) on endothelial cells and induces MAPKp38 (mitogen activated protein kinase protein 38) and NFkB (nuclear factor kappa-light-chain-enhancer of activated B cells) proinflammatory signaling (reviewed by [22] and [17]).
The alternative pathway, which is at least as old as the lectin pathway, becomes activated when MASP-3 cleaves pro-Factor D, enabling factor D to create the alternative pathway C3 convertase. In the absence of MASP-3, only thrombin possibly cleaves pro Factor D, but under circumstances of ongoing coagulation [23]. MASP-3 and Map44 also compete with MASP-1 and MASP-2 for the same binding sites on the PRMs, thus inhibiting activation of the lectin pathway [24]. MAp44, highly expressed in the heart [25], also has the ability of displacing MASPs from within the collagenous stalks of the PRMs, which may reduce myocardial tissue damage after ischemia-reperfusion injury [26–28]. Rare mutations in a highly conserved region of exon 12 of the MASP1 gene, which is exclusive of MASP-3 and encodes the serine protease domain of this protein, cause the 3MC1 (Malpuech-Michels-Mingarelli-Carnevale) syndrome, pointing to an important role in ectodermal development [29].
The possible roles of MASPs in the establishment of infections and in leprosy progression are still poorly understood. Low MASP-2 levels, as well as MASP2 polymorphisms associated with low MASP-2 production, were associated with increased susceptibility to leprosy [8]. Low MBL levels and corresponding MBL2 polymorphisms, in contrast, were associated with increased resistance [11,12], and higher FCN-3 levels were more frequent in leprosy patients than in controls [14]. It has also been suggested that complement receptor CR1 and CD91/calreticulin bind the collagenous chains of collectins and ficolins deposited on pathogens or altered cells, leading to their internalization, but that MASPs and MAps compete with this binding site, preventing this recognition [30,31]. CR1 binds opsonized M. leprae to enter the cell [32], and may use C3b and collectins/ficolins. Interestingly, we recently found polymorphisms of the CR1 gene associated with leprosy, as well as a negative correlation between the anti-inflammatory soluble CR1 and pro-inflammatory MBL levels, probably preventing inflammation [15]. All these associations were found with South Brazilian leprosy patients.
Given this context, we investigated whether MASP1 gene variants and products are associated with susceptibility to leprosy and to the different clinical forms of the disease. We aim to provide a better understanding of the immunological clinical spectrum of leprosy and of the role played by the lectin pathway in mycobacterial infections.
Materials and methods
Ethics statement
This case-control, cross-sectional study was conducted according to the Declaration of Helsinki. The local medical ethics committee of the HC-UFPR approved the study (protocol 497.079/2002–06, 218.104 and 279.970). All subjects were adults and signed a written informed consent.
Subjects and samples
We included 196 leprosy patients, of which 138 were consecutive outpatients from the Clinical Hospital of the Federal University of Paraná (HC-UFPR) and 58, inpatients from the Sanitary and Dermatologic Hospital of Paraná, both in Curitiba, Brazil. Brazilian health workers are required to enter the clinical characteristics of each leprosy patient into the “SINAN” database (Sistema de Informação de Agravos de Notificação). Each patient file contains information regarding: (1) number and location of cutaneous lesions and affected neural trunks, (2) degree of physical disability with involvement of the eyes, hands and feet, (3) bacilloscopic exam (acid-resistant bacilli screening in serous skin) and (4) histopathology results. Based on the registered clinical and histopathological features, patients were diagnosed and classified according to Ridley and Jopling criteria [33]. As the lepromatous form of leprosy is not only the most disabling, but also the most prevalent in our cohort, we sought to carry out comparisons between this patient group and the less-affected, non-lepromatous individuals. For association analyses regarding this clinical presentation, the lepromatous group comprised only patients presenting the lepromatous form of leprosy, while the non-lepromatous group comprised patients classified in all the other forms, namely tuberculoid, borderline and intermediate (those non-specified were not included). The power of the allele and haplotype analysis within the patient group was too low for detecting small effects, and results shall be taken with caution. On the other hand, within this same group, the power of the comparison of protein serum levels reached 70%, increasing our confidence in the results.
The control group comprised 214 blood donors with the same socioeconomic, ethnic and geographic background as the patients, according to information from the Hemepar and HC-UFPR blood banks. Patients and controls were defined as Euro-, Afro-Brazilians or Amerindians, based on physical characteristics and ancestry information based on the origin of first-degree relatives, collected upon patient consultations or from blood bank files. This actually means 9% sub-Saharan African and 5% average Amerindian genetic component for Euro-Brazilians, and at least 40% of African and 6% of Amerindian ancestry for Afro-Brazilians, based on the HLA allele distribution of South Brazilian populations, formerly classified in the same way. Amerindians present a very low admixture degree with European and African populations [34,35] (Table 1). Blood was collected with, or without anticoagulant ethylenediaminetetraacetic acid (EDTA) for serum collection, and DNA was extracted from peripheral blood mononuclear cells through commercial kits (Qiagen GmbH, Hilden, Germany and GFX Genomic Blood DNA Purification Kit, GE Healthcare, São Paulo, Brazil).
Table 1. Clinical and demographic description of controls and leprosy patients.
| Parameters | Controls | Patients | Exact P value |
|---|---|---|---|
|
N Age average [Min-Max] Male (%) |
214 38.17 [18–61] 116 (54.2) |
196 51.31 [18–94] 119 (60.7) |
- <0.0001 0.195 |
| Ethnical background (%)* | 0.70 | ||
| Euro-Brazilian | 176 (82.2) | 158 (80.6) | - |
| Afro-descendant Amerindian |
34 (17.3) 4 (2.1) |
36 (18.6) 2 (1.1) |
- - |
| Clinical Form (%) | |||
| Lepromatous | n.a. | 118 (60.2) | n.a. |
| Borderline | n.a. | 27 (13.7) | n.a. |
| Tuberculoid Indeterminate Non-specified |
n.a. n.a. n.a. |
18 (9.2) 10 (5.1) 23 (11.7) |
n.a. n.a. n.a. |
n: number of individuals; na.: not applicable
*: Ethnic background based on physical characteristics and ancestral information, corroborated by HLA genotyping of South-Brazilians classified in the same way (Probst et al. 2000, Braun-Prado et al. 2000).
MASP1 genotyping and haplotyping
A sequence-specific multiplex amplification method (multiplex PCR-SSP) was optimized in order to haplotype five single nucleotide polymorphisms (SNPs): rs7609662*G>A and rs13064994*C>T in intron 1 and rs72549262*G>C, rs1109452*C>T and rs850314*G>A in exon 12 within the 3’ untranslated (UTR) region (reference sequence: ENST00000337774.9). We amplified a 730 bp fragment specific for rs7609662 and rs13064994 in intron 1 and co-amplified a 365 bp fragment specific for rs72549262 and rs1109452+rs850314 (both are adjacent SNPs) in exon 12, all in a batch of four low-cost reactions, as previously described for MASP2 [8]. As a control for the amplification quality, we co-amplified a 500 bp fragment in every single reaction, corresponding to exon 8 of the Ficolin 2 gene (FCN2) by adding two generic primers (Table 2). The protocol starts with a denaturation step of 3 min at 96°C, followed by 35 cycles of 20 sec at 94°C for denaturation, 30 sec for primer annealing at variable temperatures (see below), and 30 sec DNA extension at 72C, concluding with 1 min and 30 sec at 72°C or extension. We used three different annealing temperatures according to previously published “touch-down” protocol: the 10 first cycles at 61C, followed by 10 cycles at 59°C and 15 cycles at 57°C. The haplotypes defined by these five SNPs, were identified by the presence or absence of specific bands in agarose gel, after electrophoresis.
Table 2. MASP1 sequence-specific primers and fragment size.
| Forward Primers | Reverse Primers | Fragment size |
|---|---|---|
| Intron 01 | ||
| MASP1 rs7609662_Af 5’ ATATTTGTTTCATATGTTTGAAACCA 3’ | MASP1 rs13064994_Cr 5’ TTCTTAAACCAATCTGTGGAAG 3’ | 730 bp |
| MASP1 rs7609662_Gf 5’ ATATTTGTTTCATATGTTTGAAACCG 3’ | MASP1 rs13064994_Tr 5’ TTCTTAAACCAATCTGTGGAAA 3’ | 730 bp |
| Exon 12 | ||
| MASP1 rs72549262_Cf 5’ CCCTCTCTCTTAGTGTGATC 3’ | MASP1 rs1109452_Tr 5’ CGACTAAGTCCCCATATTCA 3’ | 365 bp |
| MASP1 rs72549262_Gf 5’ CCCTCTCTCTTAGTGTGATG 3’ |
MASP1 rs1109452_Cr 5’ CGACTAAGTCCCCATATTCG 3’ MASP1 rs850314_Ar 5’ CGACTAAGTCCCCATATTT 3’ |
365 bp 366 bp |
Each primer is named after the SNP it amplifies, f: forward; r: reverse. In bold: variant nucleotides; bp: base pairs.
Using haplotype phasing through sequence-specific amplification, we identified three intron 1 haplotypes: (AC, GC, GT) and four exon 12 haplotypes (CCA, CCG, CTG and GTG). We reconstructed the whole haplotypes using a Pseudo-Bayesian Algorithm for combining intron 1 and exon 12 haplotypes, implemented in the ARLEQUIN v.3.1 software. This approach resulted in a total of twelve MASP1 haplotypes in leprosy patients and thirteen in controls. All these haplotypes were analyzed for possible associations with leprosy and with MASP-1, MASP-3 and MAp44 levels.
MASP-3 and MAp44 levels assays
Serum concentrations of MASP-3 and MAp44 were determined by time-resolved immunofluorimetric assays (TRIFMA) for 142 and 145 patients, respectively, and 116 controls, as previously described [27]. Briefly, samples were diluted in binding buffer, 40-fold for MAp44 detection and 100-fold for MASP-3, and incubated in microtiter wells coated with a monoclonal antibody. The bound protein is detected by a specific biotin-labeled monoclonal antibody, which is then subsequently detected by europium-labeled streptavidin. The provided signal is measured by time-resolved fluorometry. Four internal controls were added to each assay plate in both assays.
MASP-1 levels assay
The time-resolved immunofluorimetric assay for MASP-1 is an inhibition assay, where circulating MASP-1 in the sample inhibits the binding of an anti-MASP-1 antibody to a surface coated with a fragment of MASP-1, as previously described [36]. Briefly, diluted serum samples of 141 patients and 116 controls, 60-fold in binding buffer, were incubated with an equal volume of diluted rat anti-MASP-1 antibody for approximately an hour and then added to the coated microtiter wells. Bound rat anti-MASP-1 were detected with biotinylated rabbit anti-rat-Ig followed by europium-labeled streptavidin, where bound europium is measured by time-resolved fluorometry. Four internal controls were also added to each plate for this assay.
Statistics
Genotype, allele and haplotype frequencies were obtained by direct counting. The expectation maximization (EM) algorithm was used to calculate maximum likelihood estimates of intron 1 –exon 12 haplotype frequencies, while taking into account phase ambiguity. The hypothesis of Hardy–Weinberg equilibrium and of homogeneity between allelic distributions (exact test of population differentiation of Raymond and Rousset) was also evaluated with the ARLEQUIN software package version 3.1 (http://cmpg.unibe.ch/software/arlequin3/). We used the Wilcoxon-signed rank to define significant threshold levels and compared the distribution of protein serum concentrations between the groups, using nonparametric Mann-Whitney/Kruskal–Wallis tests (since their distribution did not pass Shapiro-Wilk normality test), with Graphpad Prism 5.01 (GraphPad Software, La Jolla, CA). The reduced model of multivariate logistic regression was used to adjust results for demographic factors; as age, sex (factors that might influence protein levels [37]) and ancestry, using STATA v.9.2 (Statacorp, TX, USA). The P values obtained with multiple comparisons in the association studies were corrected with the Benjamini-Hochberg method.
Accession numbers/ID numbers for MASP1 gene
NCBI: 5648, MIM: 600521, ENSEMBL: ENSG00000127241, hprd: 02749, UniProtKB: P48740.
Results
Protein serum levels in Southern-Brazilian patients and controls were within the range reported for a Danish population [24,27]. We found strong evidences for an association between MASP-3 and MAp44 serum levels and leprosy. We also identified a genetic association between MASP-1 and MASP-3 serum levels and MASP1 polymorphisms within haplotypes associated with increased resistance and susceptibility to leprosy. The results are described in detail below.
MASP-3 and MAp44 levels are associated with leprosy per se and lepromatous leprosy
First, we assessed possible associations of MASP-1, MASP-3 and MAp44 levels with leprosy and the different clinical forms of the disease. Leprosy patients presented lower MASP-3 levels (median 4,488 [1,722–14,634] ng/mL), than controls (median 5,575 [2,149–12,579] ng/mL) (Mann-Whitney P<0.001). In fact, the frequency of individuals with more than 5,500 ng/mL circulating MASP-3 in serum (a threshold defined with the Wilcoxon signed rank test–P<0.0004) was higher among controls: 51.7% or 60/116, compared with 31.7% or 45/142 in patients, independently of age and sex distribution (logistic regression OR = 0.51 [95%CI = 0.28–0.92] P = 0.026) (S1 Fig). Additionally, MASP-3 levels were even lower in lepromatous patients, the multibacilary form of disease presenting an exacerbated Th2 immune response. In these severely affected, often disabled patients, the median of MASP-3 levels was 4,209 [1,722–11,244] ng/mL, compared with 5,334 [2,21–14,634] ng/mL in patients with the other clinical forms (Mann-Whitney P = 0.0083). Individuals with MASP-3 levels higher than 5,500 ng/mL were also much more frequent among non-lepromatous (50% or 18/36), compared with lepromatous patients (26.8% or 26/97), independently of age and sex distribution (logistic regression OR = 0.38 [95%CI = 0.16–0.90], P = 0.028).
Similar associations were observed when analyzing MAp44 levels, but with a more conspicuous difference. Leprosy patients also presented lower MAp44 levels (median 1,715 [719–4,843] ng/mL in patients vs. median 2,330 [1,140–4,927] ng/mL in controls; Mann-Whitney P<0.0001) (Fig 1). As in the case of MASP-3, individuals with MAp44 levels higher than 2,300 ng/mL (a threshold level defined with the Wilcoxon-signed rank test – P<0.0001) were much more frequent among controls: 50.9% or 59/116, compared with 22.1% or 32/145 in patients, independently of age and sex distribution (logistic regression OR = 0.26 [95%CI = 0.14–0.49] P<0.0001) (S1 Fig). This pattern was also followed by lepromatous, compared with non-lepromatous patients: MAp44 median 1,646 [719–4,843] ng/mL vs. median 1,995 [985–4,359] ng/mL, respectively (Mann-Whitney P = 0.0021) (Fig 2). Individuals with MAp44 levels higher than 2,300 ng/mL were also much more frequent among non lepromatous (36.1% or 13/36), compared with lepromatous patients (15.5% or 15/97), again independent of age and sex distribution (logistic regression OR = 0.34 [95%CI = 0.13–0.89], P = 0.023).
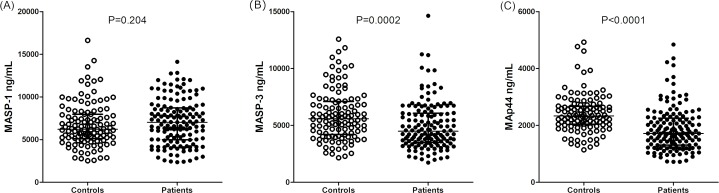
Fig 1.
MASP-1 (A), MASP-3 (B) and MAp44 (C) serum levels in controls and leprosy patients. Data shown with medians and interquartile ranges and Mann-Whitney P values. Open and closed symbols represent controls and patients, respectively.
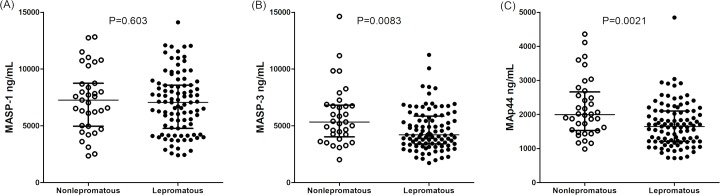
Fig 2.
MASP-1 (A), MASP-3 (B) and MAp44 (C) serum levels in non-lepromatous and lepromatous patients. Data shown with medians and interquartile ranges and Mann-Whitney P values. Open and closed symbols represent controls and patients, respectively.
In contrast, MASP-1 levels did not differ between patients and controls (median 7,036 [2,350–14,109] ng/mL vs. 6,207 [2,521–16,624] ng/mL, respectively; Mann-Whitney P = 0.173) or among the lepromatous patients and those with the other clinical forms (Mann-Whitney P = 0.603) (Figs 1 and 2).
MAp44 levels correlated significantly, but weakly, with the other two serine proteases (MASP-1: R = 0.21 in patients, Spearman P<0.05; MASP-3: R = 0.05 in patients, R = 0.36 in controls, both with Spearman P<0.0001). No correlation was found between MASP-1 and MASP-3 levels or MAp44 and MASP-1 levels in controls (S1 Fig). These results were expected, according to former reports [24] [36].
MASP1 polymorphisms and haplotypes associated with leprosy
The allele frequencies for the investigated MASP1 SNPs did not differ from Iberians (who contributed most to the Southern-Brazilian population), as well as from other Europeans, according to the 1000 Genomes project (exact test of population differentiation) (2014) (Table 3).
Table 3. Association of MASP1 variants and haplotypes with leprosy.
The intron 1 and exon 12 haplotypes were unambiguously build with sequence-specific amplification. The phase between them (symbolized by “_”) was inferred using the expectation maximization algorithm. Official SNP nomenclature is given within parenthesis for the longest cDNA, corresponding to the mRNA transcript encoding MASP-1: ENST00000337774.9. Addit: Additive association model, which tests the hypothesis that homozygosity and heterozygosity for the minor allele are associated with leprosy (either with protection or with susceptibility), but homozygosity is stronger associated, than heterozygosity. Dom: Dominant association model, which tests the hypothesis that the carrier status of the minor allele (regardless if homozygous or heterozygous) is associated with leprosy (either with protection or with susceptibility). All associations were corrected for age, which was the only demographic factor that remained associated in the reduced model of logistic regression. *: GT_GTG + GT_CTG association. q*: Benjamini-Hochberg corrected p values; ns: not significant; OR: odds ratio; CI: confidence interval.
| Variants | Iberian % (n) |
Controls % (n) |
Patients % (n) |
Lepromatous % (n) |
Others % (n) |
Model | Patients vs. Controls | |
|---|---|---|---|---|---|---|---|---|
| Total genotypes | 100 (107) | 100 (214) | 100 (196) | 100 (118) | 100 (55) | |||
| rs7609662 (c.5+2718G>A) | OR [95% CI] | P (q*) value | ||||||
| A | 13.6 (29) | 14.1 (60) | 14.6 (58) | 13.5 (32) | 13.6 (15) | ns | ns | |
| G/G | 74.8 (80) | 74.3 (159) | 72 (141) | 72.8 (86) | 74.5 (41) | ns | ns | |
| G/A | 23.4 (25) | 23.3 (50) | 27 (53) | 27.2 (32) | 23.6 (13) | ns | ns | |
| A/A | 1.9 (2) | 2.3 (5) | 1 (2) | 0 (0) | 1.8 (1) | ns | ns | |
| rs13064994 (c.6-2172C>T) | ||||||||
| T | 26.9 (49) | 28.5 (123) | 27.8 (109) | 30.1 (71) | 27.3 (30) | ns | ns | |
| C/C | 54.2 (58) | 50 (107) | 50.5 (99) | 45.7 (54) | 54.5 (30) | ns | ns | |
| C/T | 33.6 (36) | 43 (92) | 43.3 (85) | 48.3 (57) | 36.3 (20) | ns | ns | |
| T/T | 12.1 (13) | 7 (15) | 16.1 (12) | 3 (7) | 9.1 (5) | ns | ns | |
| rs72549262 (c.1304-5229C>G) | ||||||||
| G | 8.9 (19) | 11.3 (48) | 7.7 (30) | 6.8 (16) | 5.4 (6) | ns | ns | |
| C/C | 82.2 (88) | 80.8 (173) | 87.2 (171) | 84.7 (100) | 91 (50) | ns | ns | |
| C/G | 17.8 (19) | 15.8 (34) | 10.2 (20) | 10.2 (12) | 7.3 (4) | ns | ns | |
| G/G | 0(0) | 3.2 (7) | 2.5 (5) | 1.7 (2) | 1.8 (1) | ns | ns | |
| rs1109452 (c.1304-4903C>T) | ||||||||
| T | 25.2 (54) | 33.5 (143) | 33.7 (132) | 35.6 (84) | 30 (33) | ns | ns | |
| C/C | 57.9 (62) | 46.3 (99) | 44.9 (88) | 41.5 (49) | 51 (28) | ns | ns | |
| C/T | 33.6 (36) | 40.6 (87) | 42.8 (84) | 45.7 (54) | 38.2 (21) | ns | ns | |
| T/T | 8.4 (9) | 13.1 (28 | 12.2 (24) | 12.7 (15) | 10.9 (6) | ns | ns | |
| rs850314 (c.1304-4902G>A) | ||||||||
| A | 32.7 (70) | 19.9 (86) | 15.3 (60) | 13.1 (31) | 18.1 (20) | ns | ns | |
| G/G | 47.7 (51) | 64 (137) | 73 (143) | 75.4 (89) | 71 (39) | ns | ns | |
| G/A | 39.3 (42) | 32.2 (69) | 23.4 (46) | 22.9 (27) | 21.8 (12) | Dom | 0.60 [0.37–0.96] | 0.035 (0.0438) |
| A/A | 13.1 (14) | 3.7 (8) | 3.5 (7) | 1.7 (2) | 7.3 (4) | ns | ns | |
| Intron 1_Exon 12 Haplotypes | ||||||||
| GT_GTG * | 0.7 (3) | 1.8 (7) | 1.7 (4) | 0.9 (1) | Addit | 2.19 [1.18–4.03] | 0.012 (0.025) | |
| GT_CTG | 6.1 (26) | 9.2 (36) | 10.6 (25) | 9.1 (10) | Dom | 2.01 [1.05–3.83] | 0.033 (0.03) | |
| GT_CCG | 16.6 (71) | 12.2 (48) | 12.7 (30) | 11.8 (13) | Dom | 0.52 [0.32–0.86] | 0.011 (0.0188) | |
| GT_CCA | 5.1 (22) | 4.6 (18) | 5.1 (12) | 5.4 (6) | ns | ns | ||
| GC_GTG | 49.3 (40) | 5.6 (22) | 7.2 (17) | 4.5 (5) | ns | ns | ||
| GC_CTG | 13.7 (59) | 14 (55) | 13.6 (32) | 12.7 (14) | ns | ns | ||
| GC_CCA | 14.0 (60) | 9.4 (37) | 6.7 (16) | 11.8 (13) | Dom | 0.48 [0.29–0.82] | 0.008 (0.0125) | |
| GC_CCG | 20.3 (87) | 28.6 (112) | 28.8 (68) | 30 (33) | Addit | 1.70 [1.21–2.40] | 0.002 (0.0063) | |
| AC_CCA | 2.3 (10) | 1.2 (5) | 1.2 (3) | 0.9 (1) | ns | ns | ||
| AC_CTG | 0.7 (3) | 2.8 (11) | 2.5 (6) | 1.8 (2) | ns | ns | ||
| AC_GTG | 1.1 (5) | 0.2 (1) | 0 (0) | 0.9 (1) | ns | ns | ||
| AC_CCG | 9.8 (42) | 10.2 (40) | 9.7 (23) | 10 (11) | ns | ns |
We carried out association analyses with the different haplotypes formed by intron 1 and exon 12 SNPs, as well as with all 5 SNPs combined. For intron 1 variants, we identified three haplotypes: AC, GC and GT. The GC combination accounted for more than half of all intron 1 haplotypes in the investigated groups. For exon 12 variants, we identified four haplotypes: CCA, CCG, CTG and GTG. Of these, CCG was the most common, but none of the others presented less than 5% frequency. The genotypic distributions of these haplotype combinations were in Hardy and Weinberg equilibrium, except for the distribution of exon 12 haplotypes in patients (P = 0.01). Analyses with these haplotypes revealed that distribution differed between leprosy patients and controls (exact test P = 0.016), as well as between lepromatous patients and controls (exact test P = 0.023), but not between lepromatous and non-lepromatous patients. In accordance with this absence of significant difference, there was no association of MASP1 alleles/haplotypes/genotypes with the lepromatous clinical form of the disease, compared with the group containing the non-lepromatous forms. Furthermore, no associations with the disease (leprosy per se) occurred with the two variants located in intron 1. All other associations were still significant after correction for multiple comparisons (q value) and for age (the only demographic factor that remained associated with the disease, in the reduced logistic regression model).
Linkage disequilibrium between the intron 1 and exon 12 alleles resulted in a total of twelve different MASP1 haplotypes in leprosy patients and thirteen in controls, among which those with frequencies higher than 10% were GC_CCG, followed by GT_CCG, GC_CTG, GC_CCA and AC_CCG. Three of them were associated with leprosy, independently of any other demographic factor (Table 3).
Regarding the haplotypes formed by all variants analyzed in this work, the strongest association was found with the most frequent GC_CCG haplotype, which was associated with an additive (allele-dosage) susceptibility effect (OR = 1.70 [95%CI = 1.21–2.40], P<0.005). This is explained by a higher frequency of GC_CCG homozygotes and of GC_CCG heterozygotes among leprosy patients (21/196 or 10.71% and 70/196 or 35.71%), than among controls (16/214 or 7.48% and 55/214 or 25.7%), respectively. A dominant, age-dependent effect towards leprosy susceptibility was associated with carrying the less frequent GT_CTG haplotype (OR = 2.01 [95%CI = 1.06–3.83], P = 0.033). In other words, older individuals with this haplotype seem more prone to develop leprosy, if infected: there was 35/196 or 17.9% leprosy patients with GT_CTG, of which 26/35 or 74.3% with at least 40 years of age. In comparison, only 24/214 or 11.21% controls carried this haplotype, and only a third of them (8/24 or 33.3%) were 40 years of age or older. Nevertheless, the same analysis with either GT_CTG and/or another uncommon haplotype with GT in intron 1, namely GT_GTG, turned the association age-independent (OR = 2.19 [95%CI = 1.18–4.03], P = 0.012). Thus, age-dependency has a rather weak effect or may simply result from sampling bias.
In contrast, two haplotypes were associated with protection against leprosy. Among them, GC_CCA was associated with a dominant protective effect (OR = 0.48 [95%CI = 0.29–0.82], P = 0.008). This means that carriers of this haplotype were much more frequent among controls (59/214 or 27.6%), than among leprosy patients (36/196 or 18.4%). Similarly, controls presented a higher frequency of GT_CCG carriers (71/214 or 33.2%), compared with leprosy patients (43/196 or 21.9%). This haplotype was also associated with a dominant resistance effect against the disease (OR = 0.53 [95%CI = 0.32–0.86], P = 0.011) (Table 3).
MASP1 polymorphisms associated with protein serum levels
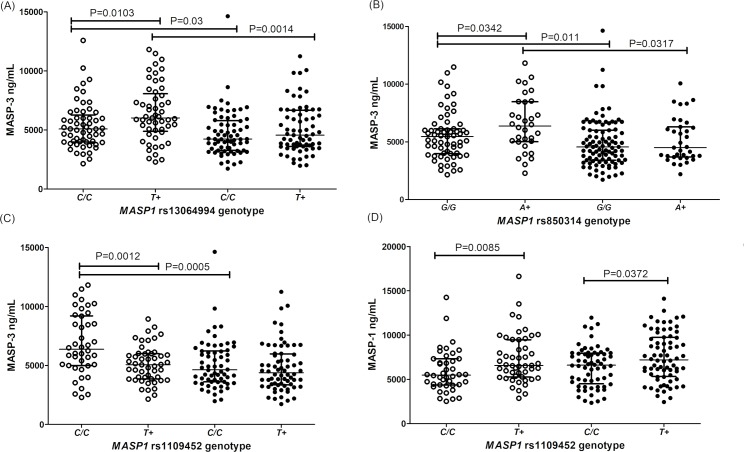
There was no association between the intron 1 rs7609662*G>A variant and MASP1 protein concentrations. Yet, the neighboring intron 1 rs13064994*C>T polymorphism was associated with MASP-3 serum concentrations: healthy carriers with the rs13064994*T variant presented higher MASP-3 levels, than C/C homozygotes (medians 6,022 [2,286–11,820] ng/mL vs. 5,086 [2,149–12,580] ng/mL, respectively, P = 0.0103). This difference disappeared among leprosy patients, whose MASP-3 concentrations reached lower levels, independent of the genotype (medians 4,557 and 4,228 ng/mL, respectively) (Fig 3A).
Fig 3. Association between variant alleles and MASP levels.
(A) rs13064994 in intron 1 and MASP-3; (B) rs850314 in exon 12 and MASP-3; (C) rs1109452 in exon 12 and MASP-3; (D) rs1109452 in exon 12 and MASP-1. Data shown with medians and interquartile ranges and Mann-Whitney P values. Open and closed symbols represent controls and patients, respectively.
Regarding the exon 12 variants, there was no association with the rs72549262 variant. However, controls with the minor rs850314*A allele of exon 12 presented higher MASP-3 levels, than G/G homozygotes (6,373 [2,286–11,820] ng/mL vs. 5,450 [2,149–11,480] ng/mL, P = 0.0342). This difference was not noticeable among patients, whose MASP-3 levels were generally lower (medians 4,500–4,554 ng/mL) and seemed no longer to be under the same genetic control (Fig 3B). In contrast, carriers of the minor rs1109452*T allele presented lower MASP-3 levels in controls, although they did not differ between healthy and diseased carriers (Fig 3C). Contrary to MASP-3 levels, MASP-1 serum concentration of rs1109452*T carriers were higher than in C/C homozygotes, independent of the disease (Fig 3D).
The adjacent exon 12 rs1109452*C and rs850314*A, as well as rs1109452*T and rs850314*G variants, occur in absolute linkage disequilibrium. The CA, CG and TG haplotype combinations did not present any association with MASP-1 and MAp44 levels (Fig 4A and 4C), although leprosy patients presented consistently lower MAp44 levels, regardless of the exon 12 genotype (Fig 4C). Healthy individuals with the CA or CG haplotype, presented higher MASP-3 concentrations than those with the TG haplotype (CA median 6,521–11,820] ng/mL and CG median 5,858 [2,286–11,820] ng/mL vs TG median 5,071 [2,149–8,941] ng/mL). In contrast to individuals with the CA and CG haplotypes, baseline levels of healthy individuals carrying TG do not differ from those with leprosy (Fig 4B).
Fig 4.
Association between haplotypes with the rs1109452 and rs850314 adjacent exon 12 variants and levels of MASP1 products, MASP-1 (A), MASP-3 (B) and MAp44 (C). Data shown with medians and interquartile ranges and Mann-Whitney P values. Open and closed symbols represent controls and patients, respectively. CA+: carriers of the rs1109452*C and rs850314*A variants. CG+: carriers of the rs1109452*C and rs850314*G variants. TG+: carriers of the rs1109452*T and rs850314*G variants. Unless if otherwise stated, comparisons were made with Mann-Whitney test.
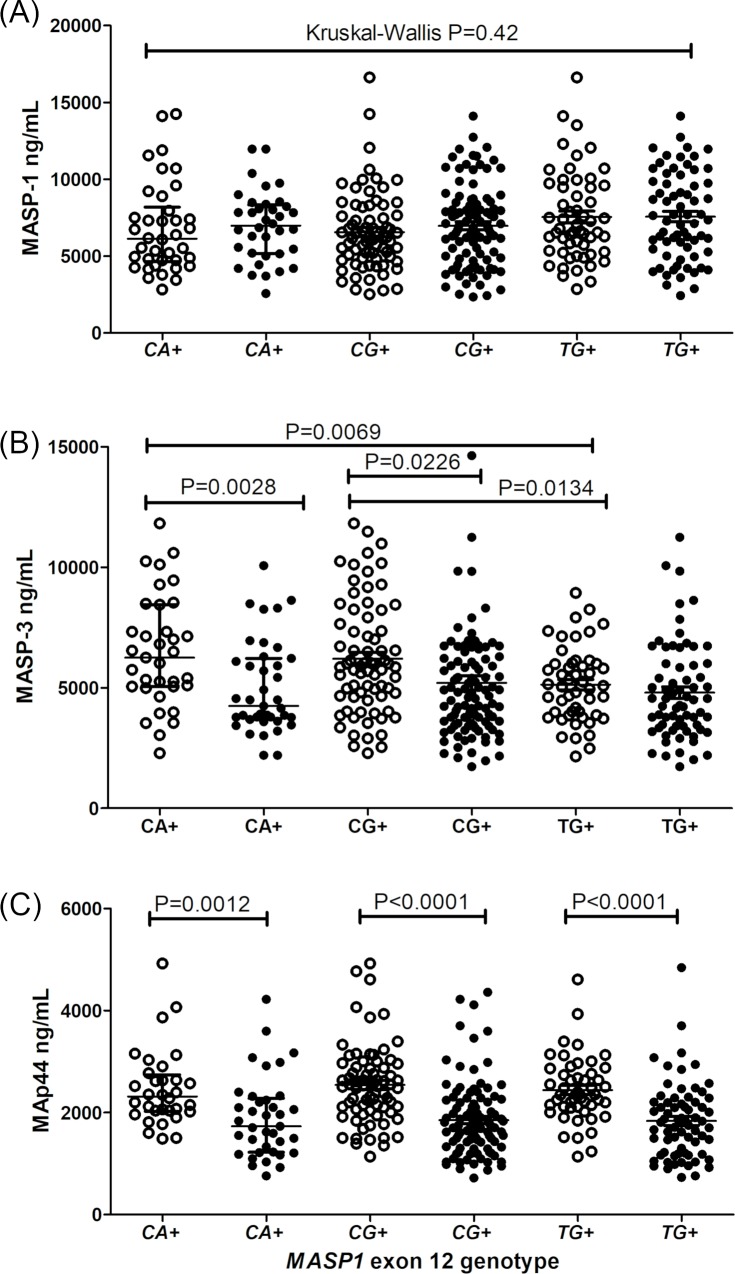
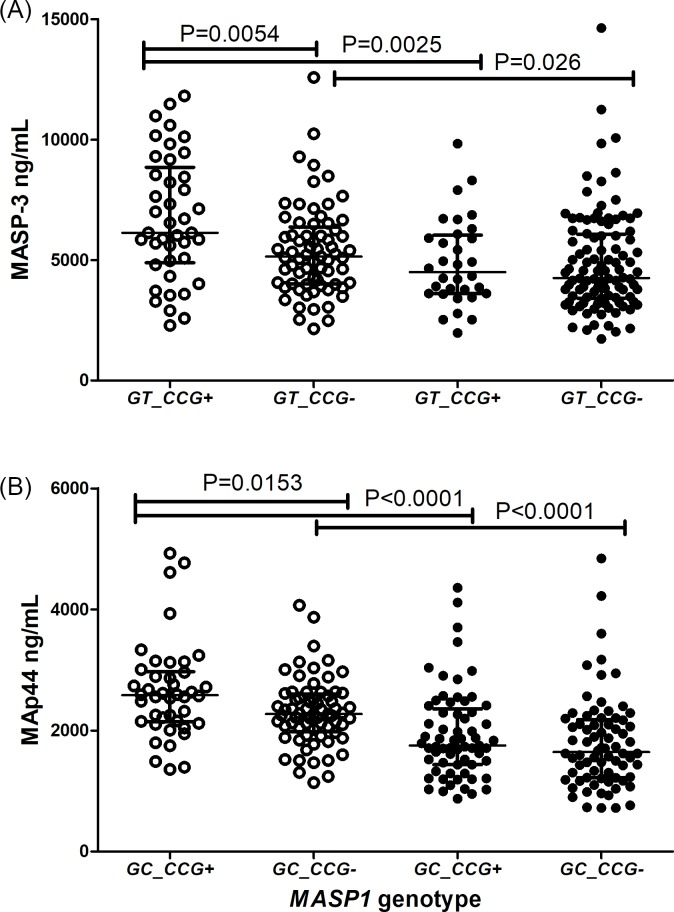
Healthy individuals carrying the GT_CCG haplotype presented higher MASP-3 levels than those without it (median: 6,131 [2,286–11,820] ng/mL vs. 5,148 [2,149–12,580] ng/mL), a difference no longer noticed among leprosy patients (Fig 5A). Similarly, controls with the GC_CCG haplotype, but not patients, presented higher MAp44 levels (median 2,581 [1,355–4,927] ng/mL vs. 2,272 [1,140–4,068] ng/mL) (Fig 5B).
Fig 5.
MASP1 haplotypes associated with (A) MASP-3 and (B) MAp44 levels. Data shown with medians and interquartile ranges and Mann-Whitney P values. Open and closed symbols represent controls and patients, respectively. + with the haplotype,—without the haplotype.
Discussion
Parasitic mycobacteria are known to usurp and efficiently evade the host defense response (reviewed by [38] & [39]). However, investigating the immune response elicited by M. leprae remains a particular challenge, due to its extreme dependence on the human host. Genetic disease association studies shed light on a wide range of aspects from the onset of infection to disease classification, by uncovering genes whose protein products may play pivotal roles in this pathology [40]. This has been the case for several genes of the lectin pathway of complement; those encoding PRMs, MBL2 [6,11] [7], FCN1 [5], FCN2 [9] and FCN3 [14], the serine protease MASP2 [8] and the MBL receptor encoded by CR1 [15]. The evaluation of complement protein levels adds highly relevant information to this picture, as an indirect measure of gene expression, complement activation and consumption. Since the seventies, these measurements have been done for leprosy disease [41] [12,13], with results currently supported by transcriptome studies [42]. In the present investigation, we finally added MASP1 polymorphisms and protein products, as one important piece of the initiation complexes of the lectin pathway to the association of complement with leprosy disease. In fact, we carried out the analyses presented in this work with the same patient cohort from a Brazilian population [5,9,11,14,15].
To understand the possible roles of MASP1 products in the disease, it is important to keep in mind two prevailing hypotheses that may explain the role of complement proteins in leprosy disease. Firstly, they increase infection success by improving opsonization and phagocytosis of M. leprae by the host macrophage cells. Secondly, they increase inflammation after the disease is established, leading to more severe tissue damage.
Regarding the first hypothesis, it may be argued that any variant that reduces the rate of opsonin deposition would be protective, whereas any variant that increases opsonization would enhance susceptibility. According to this, one would expect that high MASP-1 levels would aid M. leprae’s entrance into host cells, whereas high MASP-3/MAp44 levels would block activation of the lectin pathway and reduce phagocytosis of the bacteria (although MASP-3 may also activate the alternative pathway). In fact, higher MASP-3 and MAp44 levels were characteristic for healthy individuals, although the expected effect was not seen for MASP-1 (Fig 6). With respect to the second hypothesis, it is expected that variants that reduce complement activation would (again) play a protective role. Indeed, we found a clear-cut difference between patients, with higher MASP-3/MAp44 levels more prevalent among those, less severely affected. Since it is known from former studies that Dapsone and Clofazimine treatment (used by the patients in this study) does not interfere with complement availability and function [43,44], it may be assumed that lower MASP-3 and MAp44 levels among patients, especially among those with the most severe lepromatous condition, are genetically determined (Fig 6).
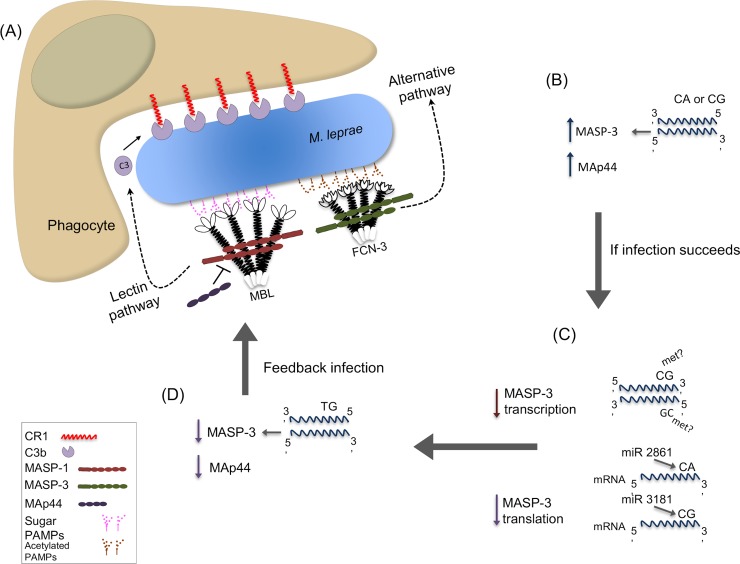
Fig 6. Proposed roles for MASP1 products and polymorphisms in susceptibility to M. leprae infection.
(A) Collectins (e.g. MBL) or ficolins (e.g. FCN-3) recognize pathogen-associated molecular patterns (PAMPs), composed of sugar/acetylated groups on M. leprae. MASP-2 (not depicted in this image) and MASP-1 homodimers complexed with them activate the lectin pathway of complement, whereas MASP-3 may activate the alternative pathway. Both pathways lead to C3b-opsonization and CR1-mediated internalization of the pathogen. (B) Healthy individuals with rs1109452 and rs850314 CA or CG haplotypes express higher MASP-3 levels. Higher MASP-3 and MAp44 levels were also associated with resistance against the disease. (C) CpG methylation at the CG haplotype in exon 12 may impair mRNA transcription, spliceosome assembly and mRNA processing. Reduced MASP-3 levels may also result from the differential recognition of CA and CG haplotypes by miRNAs (miR-2861 and miR-3181, respectively). (D) Individuals with TG haplotypes present lower baseline MASP-3 levels. Lower MASP-3 and MAp44 levels seem to predispose to the infection, possibly by optimizing opsonin coverage of the parasite.
There are numerous polymorphisms in the MASP1 gene that may interfere with gene expression, some of which had been formerly investigated by others [45] [46]. We chose to investigate two SNPs located in a regulatory region of intron 1, which may interfere with the production of all three MASP1 proteins, and three in exon 12, which is exclusive of MASP-3 and may uniquely affect the expression level of this protein. None of them had been previously investigated.
The rs7609662*A in intron 1 is associated with higher MASP1 mRNA levels in several tissues (https://gtexportal.org/home/snp/rs7609662), but we did not identify this effect at the protein level. The rs13064994*T had the opposite effect (https://gtexportal.org/home/snp/rs13064994) on MASP1 mRNA expression. We found an association of this allele with higher MASP-3 protein levels, but only in healthy individuals. The absence of a clear correlation between mRNA levels and protein concentration in serum is not unexpected, since former analyses did not consider different MASP1 transcripts, and stability of mRNA in cytoplasm may be greatly affected by regulatory mechanisms that were not accounted for in previous transcriptomic analyses.
All exon 12 variants (rs72549262*G>C, rs1109452*C>T and rs850314*G>A) are located within the 3’ untranslated region. Those two most downstream (rs1109452 and rs850314) are adjacent to each other, and CG represents the most ancestral combination. Thus, the minor alleles rs1109452*T and rs850314*A disrupt a 5’CpG3’ site (where “p” means the phosphodiester bond between rs1109452*C and rs850314*G). The cytosine of this CpG site was found methylated in the brain [47], but not in cell lines from liver and female reproductive tissue, where MASP-3 mRNA production is higher (https://gtexportal.org/home/gene/MASP1). DNA methylation in alternatively spliced exons may modulate exon inclusion [48].
Furthermore, the CA and CG combinations are miRNA targets, as predicted in silico using targetScan7.1, and may reduce MASP-3 translation. Thus, one would expect that any nucleotide substitution at these loci would modify gene expression, depending on specific regulatory requirements of the cell type, developmental stage, physiological and immunological responses. In fact, both adjacent polymorphisms were associated with MASP-3 (in the case of rs1109452, even MASP-1) levels. However, the predicted down-regulating effects either of CpG methylation and/or CA/CG miRNA binding on MASP-3 levels, were restricted to leprosy patients. In the disease, MASP-3 levels of CA or CG carriers dropped to the same concentration found in TG carriers, who presented the lowest MASP-3 levels, independent of the disease. Interestingly, among the miRNAs predicted to recognize these polymorphic sites, none bind TG, but miR-3181 preferentially recognizes CG and miR-2861, CA. Both are expressed in the liver [49], with miR-2861 being up-regulated by interleukin 6 [50], a proinflammatory cytokine with a pivotal role in leprosy disease [51]. It is thus conceivable that these regulatory mechanisms operate after disease establishment and activation of the acute phase response (Fig 6).
Refining the association analysis to the haplotype level, allowed us to identify the GT_CCG and GC_CCA haplotypes (containing the previously mentioned rs850314*A variant) associated not only with higher MASP-3 levels, but also with higher protection against the disease. Higher MASP-3 levels may avoid initiation of bacterial colonization due to competition with MASP-1 and MASP-2 for binding sites of recognition molecules—blocking the lectin pathway, and/or by competition with binding sites on complement receptors, blocking phagocytosis.
Yet the GC_CCG haplotype, associated with leprosy susceptibility, was associated with higher MAp44 serum concentrations. In contrast with MASP-3, however, MAp44 serum levels did not associate with the investigated SNPs, which may suggest other causal variants in linkage disequilibrium with GC_CCG, not investigated in this study. In fact, Ammitzboll et al. (2013) [45] list several variants that may modulate MAp44 levels. Furthermore, other factors than those regulating MASP-3 may fit in the present scenario, where MAp44 levels are higher in controls, compared to patients, and in non-lepromatous patients, compared to the more severely affected lepromatous patients.
Beside GC_CCG, the haplotypes GT_CTG and GT_GTG also present at least an additive effect increasing almost twice the susceptibility to the disease. They were not associated with protein levels, although harboring the rs1109452*T polymorphism, found associated with higher MASP-1 and lower MAp44 levels. Thus, protein levels shall not be held solely responsible for the association of MASP1 products with the disease. Beside the pleiotropic nature of the MASP1 gene itself, the investigated polymorphisms may have effects far beyond those affecting MASP1. Other variants linked with those that compose the associated haplotypes, may present epistatic and/or unsuspected pleiotropic effects that affect susceptibility to the disease. In fact, the variants investigated in this study have been recently associated with expression levels of neighboring genes as the ribosomal protein-encoding gene RPL39L and the odorant receptor transporters RTP1, RTP3 and RTP4 (https://gtexportal.org/home/gene/MASP1 and Immunpop browser). Among them, RTP4 is strongly up-regulated by interferon I, a cytokine known to suppress an adequate cellular response driven by interferon type II against M. leprae [52].
In summary, our results show associations of MASP-3 and MAp44 levels with leprosy, as well as with polymorphisms that regulate the levels of these two proteins in serum. Taken together, these results indicate that MASP-3/MAp44 blockage of the lectin pathway may not be the only explanation for resistance to leprosy infection, since expression levels of neighboring genes may be regulated by noncoding polymorphisms investigated in this study. Although interpreting the evidence is not straightforward, it certainly fosters more investigations on the role played by MASP1 products in the resistance against mycobacterial infections and its more severe forms. In particular, MASP-3 and MAp44 may be evaluated as new therapeutic agents against leprosy infection and against polarization to lepromatous disease. These observations place the MASP1 gene and its products in the cross-talk of innate and adaptive immune responses to leprosy infection, its clinical aspects and susceptibility, thus shedding more light on how the complement system may be involved with mycobacterial infections.
Supporting information
Correlations between MASP-1, MASP-3 and MAp44 serum levels in leprosy patients (A-B) and healthy controls (C-D). Linear regression fit, P and R values are shown.
(TIF)
n: number of individuals; *: mean protein levels in ug/mL showing: median[IQR] **Levels confering protection against Leprosy infection. Within brackets: minimum and maximal values. a: Patients presenting all other forms except Lepromatous and Non-specified
(TIF)
(XLSX)
Acknowledgments
We deeply thank all patients and controls that volunteered in this study. We also want to thank Sandra J. dos Santos Catarino, Annette G. Hansen and Lisbeth Jensen for all technical guidance and contributions to this work.
Data Availability
All relevant data are included within the paper and its Supporting Files. The genotyping protocols can be found at: https://www.protocols.io/view/masp1-multiplex-pcr-ssp-rs7609662-rs13064994-rs725-27kghkw. The accession numbers and their associated databases are: NCBI: 5648, MIM: 600521, ENSEMBL: ENSG00000127241, hprd: 02749, UniProtKB: P48740.
Funding Statement
This work was supported by the 01/2007 and 518/2010 PRODOC grants of CAPES (Coordenação de Aperfeiçoamento de Pessoal Superior, http://www.capes.gov.br/bolsas/bolsas-no-pais/prodoc) and by the 034/2008 CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico, http://www.cnpq.br/web/guest/bolsas-e-auxilios) and Fundação Araucária (http://www.fappr.pr.gov.br/) grants for ABW Boldt and IJT Messias-Reason. Dr. Thiel and Dr. Jensenius were funded by the Danish Research Council and the Novo Nordisk Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Hellen Weinschutz Mendes had a MSc scholarship from Coordenação de Aperfeiçoamento de Pessoal Superior (CAPES).
References
- 1.WHO, Global Leprosy Strategy. Weekly epidemiological record. 2018; 445–456. https://apps.who.int/iris/bitstream/handle/10665/274289/WER9335.pdf?ua=1 [Google Scholar]
- 2.Fava VM, Sauer MD, Schurr E. Genetics of leprosy: today and beyond. Hum Genet. Springer Berlin Heidelberg; 2019; 10.1007/s00439-019-02087-5 [DOI] [PubMed] [Google Scholar]
- 3.Endo Y, Takahashi M, Nakao M, Saiga H, Sekine H, Matsushita M, et al. Two lineages of mannose-binding lectin-associated serine protease (MASP) in vertebrates. J Immunol. 1998;161: 4924–30. http://www.jimmunol.org/content/161/9/4924.full [PubMed] [Google Scholar]
- 4.Garred P, Harboe M, Oettinger T, Koch C, Svejgaard A. Dual role of mannan-binding protein in infections: another case of heterosis? Eur J Immunogenet. 1994;21: 125–31. http://www.ncbi.nlm.nih.gov/pubmed/9098426 10.1111/j.1744-313x.1994.tb00183.x [DOI] [PubMed] [Google Scholar]
- 5.Boldt ABW, Sanchez MIN, Stahlke ERS, Steffensen R, Thiel S, Jensenius JC, et al. Susceptibility to Leprosy is Associated with M-ficolin Polymorphisms. J Clin Immunol. 2013;33: 210–219. 10.1007/s10875-012-9770-4 [DOI] [PubMed] [Google Scholar]
- 6.Zhang D-F, Huang X-Q, Wang D, Li Y-Y, Yao Y-G. Genetic variants of complement genes Ficolin-2, Mannose-binding lectin and Complement factor H are associated with leprosy in Han Chinese from Southwest China. Hum Genet. 2013;132: 629–640. 10.1007/s00439-013-1273-8 [DOI] [PubMed] [Google Scholar]
- 7.Cardona-Pemberthy V, Rendón M, Beltrán JC, Soto-Ospina A, Muñoz-Gomez A, Araque-Marín P, et al. Genetic variants, structural, and functional changes of Myelin Protein Zero and Mannose-Binding Lectin 2 protein involved in immune response and its allelic transmission in families of patients with leprosy in Colombia. Infect Genet Evol. 2018;61: 215–223. 10.1016/j.meegid.2018.04.002 [DOI] [PubMed] [Google Scholar]
- 8.Boldt ABW, Goeldner I, Stahlke ERS, Thiel S, Jensenius JC, de Messias-Reason IJT. Leprosy association with low MASP-2 levels generated by MASP2 haplotypes and polymorphisms flanking MAp19 exon 5. PLoS One. 2013;8: e69054 10.1371/journal.pone.0069054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Messias‐Reason I, Kremsner PG, Kun JFJ. Functional Haplotypes That Produce Normal Ficolin‐2 Levels Protect against Clinical Leprosy. J Infect Dis. 2009;199: 801–804. 10.1086/597070 [DOI] [PubMed] [Google Scholar]
- 10.Monot M, Honoré N, Garnier T, Zidane N, Sherafi D, Paniz-Mondolfi A, et al. Comparative genomic and phylogeographic analysis of Mycobacterium leprae. Nat Genet. 2009;41: 1282–9. 10.1038/ng.477 [DOI] [PubMed] [Google Scholar]
- 11.de Messias-Reason IJ, Boldt ABW, Moraes Braga AC, Von Rosen Seeling Stahlke E, Dornelles L, Pereira-Ferrari L, et al. The association between mannan-binding lectin gene polymorphism and clinical leprosy: new insight into an old paradigm. J Infect Dis. 2007;196: 1379–85. 10.1086/521627 [DOI] [PubMed] [Google Scholar]
- 12.Dornelles LN, Pereira-Ferrari L, Messias-Reason I. Mannan-binding lectin plasma levels in leprosy: deficiency confers protection against the lepromatous but not the tuberculoid forms. Clin Exp Immunol. 2006;145: 463–8. 10.1111/j.1365-2249.2006.03161.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gomes GI, Nahn EP, Santos RK, Da Silva WD, Kipnis TL. The functional state of the complement system in leprosy. Am J Trop Med Hyg. 2008;78: 605–10. http://www.ncbi.nlm.nih.gov/pubmed/18385356 [PubMed] [Google Scholar]
- 14.Andrade FA, Beltrame MH, Bumiller-Bini V, Boslooper Gonçalves L, Beate A, Boldt W, et al. Association of a new FCN3 haplotype with high ficolin-3 levels in leprosy. 2017; 10.1371/journal.pntd.0005409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Canalli Kretzschmar G, Caroline Oliveira L, Mitsunori Nisihara R, Velavan TP, rvio Tú lio Stinghen S, S Stahlke ER, et al. Complement receptor 1 (CR1, CD35) association with susceptibility to leprosy. 2018; 10.1371/journal.pntd.0006705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garred P, Genster N, Pilely K, Rosbjerg A, Jie Y, Skjoedt MM. A journey through the lectin pathway of complement—MBL and beyond. 2016;274: 74–97. 10.1111/imr.12468 [DOI] [PubMed] [Google Scholar]
- 17.Boldt ABW., Boschmann SE., Catarino SJ., Andrade FA. de Messias-Reason IJT. Encyclopedia of Signaling Molecules In: Choi S, editor. Encyclopedia of Signaling Molecules. Springer; 2016. pp. 2972–89. 10.1007/978-1-4614-6438-9 [DOI] [Google Scholar]
- 18.Endo Yuichi, Nonaka Masaru, Saiga Hidetoshi, Kakinuma Yuji, Matsushita Akiko, Minoru Takahashi. Origin of Mannose-Binding Lectin-Associated Serine Protease (MASP)-1 and MASP-3 Involved in the Lectin Complement Pathway Traced Back to the Invertebrate, Amphioxus. J Immunol. 2014;170: 4701–4707. 10.4049/jimmunol.170.9.4701 [DOI] [PubMed] [Google Scholar]
- 19.Krarup A, Gulla KC, Gál P, Hajela K, Sim RB. The action of MBL-associated serine protease 1 (MASP1) on factor XIII and fibrinogen. Biochim Biophys Acta— 2008;1784: 1294–1300. 10.1016/j.bbapap.2008.03.020 [DOI] [PubMed] [Google Scholar]
- 20.Schroeder V, Hess K, Dobó J, Ajjan R, Phoenix F, Gál P. Effects of MASP-1 of the Complement System on Activation of Coagulation Factors and Plasma Clot Formation. PLoS One. 2012;7: e35690 10.1371/journal.pone.0035690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jozsef Dobó J, zs Major B, Kékesi KA, Szabó I, rton Megyeri M, Hajela K, et al. Cleavage of Kininogen and Subsequent Bradykinin Release by the Complement Component: Mannose-Binding Lectin-Associated Serine Protease (MASP)-1. 2011; 10.1371/journal.pone.0020036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dobó J, Pál G, Cervenak L, Gál P. The emerging roles of mannose-binding lectin-associated serine proteases (MASPs) in the lectin pathway of complement and beyond. Immunol Rev. 2016;274: 98–111. 10.1111/imr.12460 [DOI] [PubMed] [Google Scholar]
- 23.Dobó J, Schroeder V, Jenny L, Cervenak L, Závodszky P, Gál P. Multiple roles of complement MASP-1 at the interface of innate immune response and coagulation. Mol Immunol. 2014; 1–10. 10.1016/j.molimm.2014.05.013 [DOI] [PubMed] [Google Scholar]
- 24.Skjoedt M-O, Palarasah Y, Munthe-Fog L, Jie Ma Y, Weiss G, Skjodt K, et al. MBL-associated serine protease-3 circulates in high serum concentrations predominantly in complex with Ficolin-3 and regulates Ficolin-3 mediated complement activation. Immunobiology. 2010;215: 921–31. 10.1016/j.imbio.2009.10.006 [DOI] [PubMed] [Google Scholar]
- 25.Skjoedt M-O, Hummelshoj T, Palarasah Y, Honore C, Koch C, Skjodt K, et al. A novel mannose-binding lectin/ficolin-associated protein is highly expressed in heart and skeletal muscle tissues and inhibits complement activation. J Biol Chem. 2010;285: 8234–43. 10.1074/jbc.M109.065805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Degn SE, Hansen AG, Steffensen R, Jacobsen C, Jensenius JC, Thiel S. MAp44, a human protein associated with pattern recognition molecules of the complement system and regulating the lectin pathway of complement activation. J Immunol. 2009;183: 7371–8. 10.4049/jimmunol.0902388 [DOI] [PubMed] [Google Scholar]
- 27.Degn SE, Jensen L, Gál P, Dobó J, Holmvad SH, Jensenius JC, et al. Biological variations of MASP-3 and MAp44, two splice products of the MASP1 gene involved in regulation of the complement system. J Immunol Methods. 2010;361: 37–50. 10.1016/j.jim.2010.07.006 [DOI] [PubMed] [Google Scholar]
- 28.Degn SE, Thiel S. Humoral Pattern Recognition and the Complement System. 2013; 181–193. 10.1111/sji.12070 [DOI] [PubMed] [Google Scholar]
- 29.Rooryck C, Diaz-font A, Osborn DPS, Chabchoub E, Shamseldin H, Kenny J, et al. Europe PMC Funders Group Mutations in the lectin complement pathway genes COLEC11 and MASP1 cause 3MC syndrome. 2011;43: 197–203. 10.1038/ng.757.Mutations [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacquet M, Lacroix M, Ancelet S, Gout E, Gaboriaud C, Thielens NM, et al. Deciphering Complement Receptor Type 1 Interactions with Recognition Proteins of the Lectin Complement Pathway. J Immunol. 2013;190: 3721–3731. 10.4049/jimmunol.1202451 [DOI] [PubMed] [Google Scholar]
- 31.Duus K, Thielens NM, Lacroix M, Tacnet P, Frachet P, Holmskov U, et al. CD91 interacts with mannan-binding lectin (MBL) through the MBL-associated serine protease-binding site. FEBS J. 2010;277: 4956–4964. 10.1111/j.1742-4658.2010.07901.x [DOI] [PubMed] [Google Scholar]
- 32.Schlesinger LS, Horwitz MA. Phenolic glycolipid-1 of Mycobacterium leprae binds complement component C3 in serum and mediates phagocytosis by human monocytes. J Exp Med. 1991;174: 1031–8. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2118995&tool=pmcentrez&rendertype=abstract 10.1084/jem.174.5.1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ridley DS, Jopling WH. Classification of leprosy according to immunity. A five-group system. Int J Lepr Other Mycobact Dis. 1966. 34: 255–73. http://www.ncbi.nlm.nih.gov/pubmed/5950347 [PubMed] [Google Scholar]
- 34.Braun-Prado K, Vieira Mion AL, Farah Pereira N, Culpi L, Petzl-Erler ML. HLA class I polymorphism, as characterised by PCR-SSOP, in a Brazilian exogamic population. Tissue Antigens. 2000;56: 417–27. http://www.ncbi.nlm.nih.gov/pubmed/11144289 10.1034/j.1399-0039.2000.560504.x [DOI] [PubMed] [Google Scholar]
- 35.Probst CM, Bompeixe EP, Pereira NF, de O Dalalio MM, Visentainer JE, Tsuneto LT, et al. HLA polymorphism and evaluation of European, African, and Amerindian contribution to the white and mulatto populations from Paraná, Brazil. Hum Biol. 2000;72: 597–617. http://www.ncbi.nlm.nih.gov/pubmed/11048789 [PubMed] [Google Scholar]
- 36.Thiel S, Jensen L, Degn SE, Nielsen HJ, Gál P, Dobó J, et al. Mannan-binding lectin (MBL)-associated serine protease-1 (MASP-1), a serine protease associated with humoral pattern-recognition molecules: normal and acute-phase levels in serum and stoichiometry of lectin pathway components. Clin Exp Immunol. 2012;169: 38–48. 10.1111/j.1365-2249.2012.04584.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Troldborg A, Hansen A, Hansen SWK, Jensenius JC, Stengaard-Pedersen K, Thiel S. Lectin complement pathway proteins in healthy individuals. Clin Exp Immunol. 2017;188: 138–147. 10.1111/cei.12909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ottenhoff THM. New pathways of protective and pathological host defense to mycobacteria. Trends Microbiol. 2012;20: 419–428. 10.1016/j.tim.2012.06.002 [DOI] [PubMed] [Google Scholar]
- 39.Ernst JD. The immunological life cycle of tuberculosis. Nat Rev Immunol. 2012;12: 581–591. 10.1038/nri3259 [DOI] [PubMed] [Google Scholar]
- 40.Cambri G, Mira MT. Genetic susceptibility to leprosy-from classic immune-related candidate genes to hypothesis-free, whole genome approaches. Front Immunol. 2018;9: 1–9. 10.3389/fimmu.2018.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Srivastava LM, Agarwal DP, Benkmann HG, Goedde HW. Biochemical, immunological and genetic studies in leprosy. III. Genetic polymorphism of C3 and immunoglobulin profile in leprosy patients, healthy family members and controls. Tropenmed Parasitol. 1975;26: 426–30. http://www.ncbi.nlm.nih.gov/pubmed/1216330 [PubMed] [Google Scholar]
- 42.Amorim FM, Nobre ML, Nascimento LS, Miranda AM, Monteiro GRG, Freire-Neto FP, et al. Differential immunoglobulin and complement levels in leprosy prior to development of reversal reaction and erythema nodosum leprosum. Lockwood DNJ, editor. PLoS Negl Trop Dis. 2019;13: e0007089 10.1371/journal.pntd.0007089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sahu A, Saha K, Kashyap A, Chakrabarty AK. Interaction of anti-leprosy drugs with the rat serum complement system. Immunopharmacology. 1988. 15: 143–50. http://www.ncbi.nlm.nih.gov/pubmed/3134310 10.1016/0162-3109(88)90025-2 [DOI] [PubMed] [Google Scholar]
- 44.Kashyap A, Sehgal VN, Sahu A, Saha K. Anti-leprosy drugs inhibit the complement-mediated solubilization of pre-formed immune complexes in vitro. Int J Immunopharmacol. 1992;14: 269–73. http://www.ncbi.nlm.nih.gov/pubmed/1624226 10.1016/0192-0561(92)90039-n [DOI] [PubMed] [Google Scholar]
- 45.Ammitzbøll C, Steffensen R, Nielsen H. Polymorphisms in the MASP1 gene are associated with serum levels of MASP-1, MASP-3, and MAp44. PLoS One. 2013;8: e73317 10.1371/journal.pone.0073317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ingels C, Vanhorebeek I, Steffensen R, Derese I, Jensen L, Wouters PJ, et al. Lectin pathway of complement activation and relation with clinical complications in critically ill children. Pediatr Res. 2014;75: 99–108. 10.1038/pr.2013.180 [DOI] [PubMed] [Google Scholar]
- 47.Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D’souza C, Fouse SD, et al. Conserved Role of Intragenic DNA Methylation in Regulating Alternative Promoters. Nature. 2010; 466: 253–7 10.1038/nature09165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lev Maor G, Yearim A, Ast G. The alternative role of DNA methylation in splicing regulation. Trends Genet. 2015;31: 274–80. 10.1016/j.tig.2015.03.002 [DOI] [PubMed] [Google Scholar]
- 49.Ferguson DC, Blanco JG. Regulation of the Human Fc-Neonatal Receptor alpha-Chain Gene FCGRT by MicroRNA-3181. Pharm Res. 2018; 35: 15 10.1007/s11095-017-2294-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kirchmeyer M, Servais FA, Hamdorf M, Nazarov P V, Ginolhac A, Halder R, et al. Cytokine-mediated modulation of the hepatic miRNome: miR-146b-5p is an IL-6-inducible miRNA with multiple targets. J Leukoc Biol. 2018;104: 987–1002. 10.1002/JLB.MA1217-499RR [DOI] [PubMed] [Google Scholar]
- 51.Sales-Marques C, Chester Cardoso C, Elena Alvarado-Arnez L, Illaramendi X, Maria Sales A, de Andréa Hacker M, et al. Genetic polymorphisms of the IL6 and NOD2 genes are risk factors for inflammatory reactions in leprosy. 2017; 10.1371/journal.pntd.0005754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, et al. A diverse array of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472: 481–485. 10.1038/nature09907 [DOI] [PMC free article] [PubMed] [Google Scholar]