Figure 4. Remodeling defects are independent of the source of DSB formation.
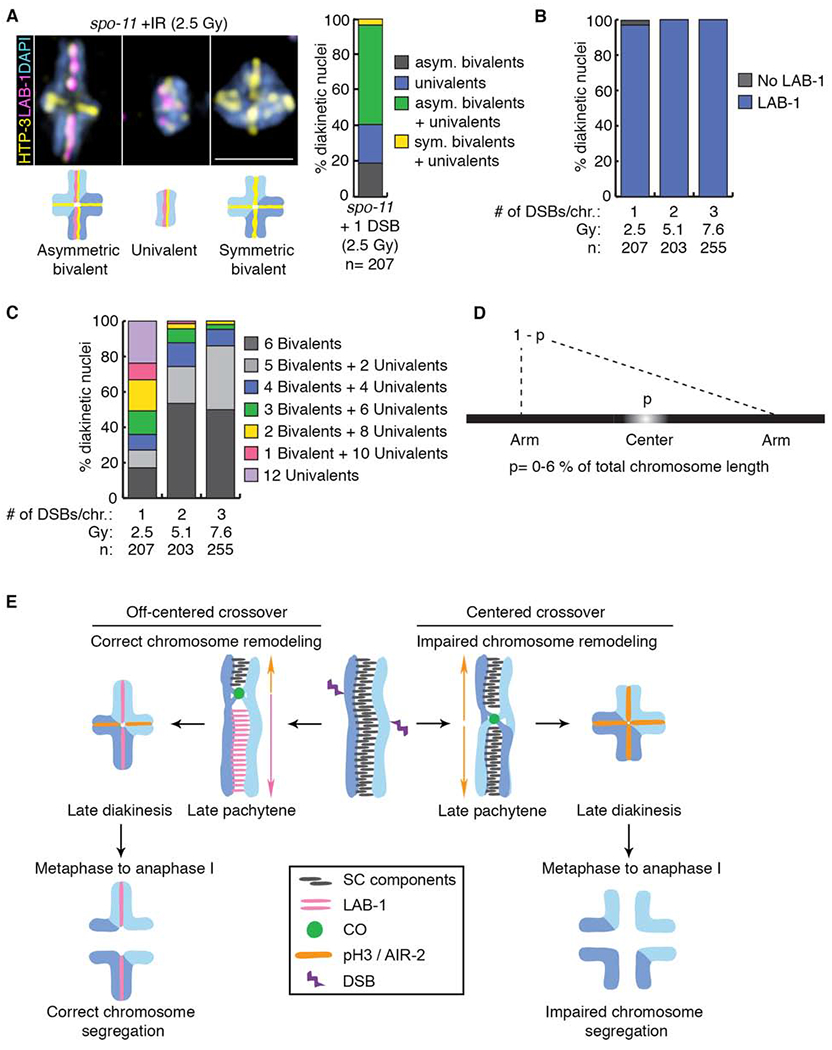
(A) Representative DAPI-stained bodies (blue) showing the localization of HTP-3 (yellow) and LAB-1 (magenta) observed in diakinesis nuclei of spo-11 mutants subjected to γ-IR doses producing 1, 2 or 3 DSBs per chromosome pair. Illustrations are shown below the immunofluorescence images. Bar, 2 μm. Histogram depicting the categories of bivalent/univalent configurations observed at diakinesis after inducing 1 DSB per homolog pair by γ-IR (2.5 Gy), n indicates the number of diakinesis nuclei examined. (B) Quantification of LAB-1 localization in diakinesis nuclei following exposure to indicated γ-IR doses. (C) Histogram showing quantification of the observed number of bivalents and/or univalents at different γ-IR doses. (D) Mathematical modeling used to determine the length of the center region leading to chromosome remodeling defects upon CO formation. (E) Model for how CO position influences late prophase chromosome remodeling and subsequent chromosome segregation. DSBs occur throughout the length of the chromosomes (up to 10 per homolog pair in C. elegans [37]), but only 1 introduced at an off-centered position is preferentially used for CO formation leading to correct chromosome remodeling resulting in asymmetric bivalents and accurate chromosome segregation. In contrast, COs occurring at the center of the chromosomes result in impaired chromosome remodeling resulting in symmetric bivalents. This chromosome configuration lacks LAB-1 localization and this in turn fails to restrict AIR-2/pH3 to a single chromosome axis resulting in premature loss of sister chromatid cohesion and errors in chromosome segregation. See also Figure S4 and Data S5.
