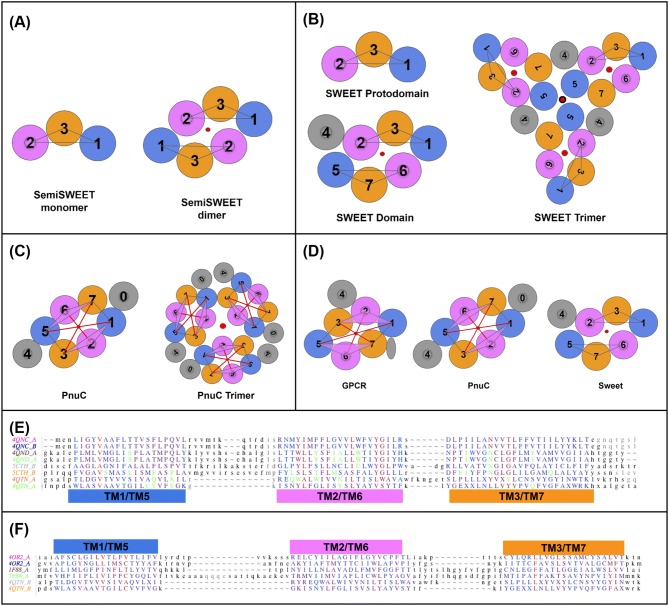
Fig. 2.
SemiSWEET, SWEET, PnuC and GPCRs. a SemiSWEET 3TMH monomer and dimer. b SWEET 3TMH protodomain and a 7TMH SWEET domain formed from 3TMH protodomains (left) and SWEET trimer (right). c PnuC 7/8TMH domain (left; see text) and trimer (right). d GPCR vs. PnuC vs. SWEET—comparison of topologies. e Structure-based sequence alignments. SemiSWEET/SWEET/PnuC—structure-based sequence alignment of 3TMH from SemiSWEET dimer (4QNC; 4QND), SWEET (5CTH), PnuC (4QTN). Ligand binding residues are in green, conserved/similar residues are in red. Structural alignment of TMHs of PnuC (pdbid 4QTN) to SemiSWEET (4QNC) and SWEET (5CTH) protodomains gives an RMSD of 6.5 Å. So, PnuC does not appear to be a structural homolog of SWEET at the protodomain level. However, from the very simple schematic representation, one can see that TMH167 and TM523 match the SWEET/SemiSWEET protodomains. If we ignore TM1 and TM5 in PnuC, the RMSD on other 2 helices goes down to 1.71 Å and 2.47 Å, respectively. The RMSD between PnuC's 3-helix combination 167 and SemiSWEET 3TMH is 1.87 Å, suggesting a structural homology and possible evolutionary link (see text). f GPCR vs. PnuC—structure-based sequence alignment of 3TMH from a class C GPCR (4OR2) vs. a class A GPCR (Rhodopsin) (1F88) vs. PnuC (4QTN). The RMSD values of multiple protodomains aligned vs. the first one (4OR2-1) are 3.63, 2.12, 3.29, 3.29, 3.34 Å for 4OR2-2, 1F88-1&2, 4QTN-1&2, respectively. However, if one excludes TM2 from the structural alignment the RMSD is now 2.09, 1.17, 2.77, 2.61, 2.79 Å, respectively