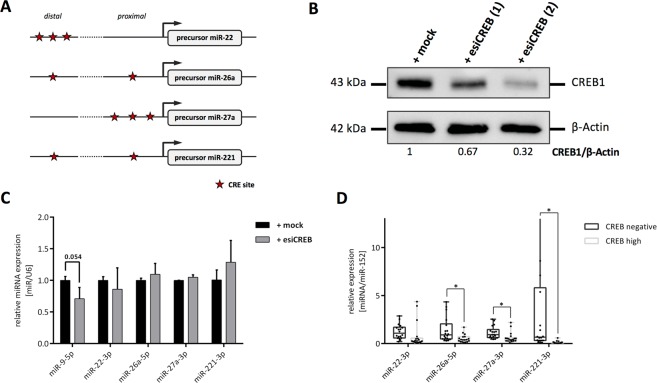
Figure 5.
Correlation of CREB1 and newly identified CREB1-specific miRNAs. (A) In silico prediction of miRNA promoter and identification of CREB1 responsive elements (CRE sites, red asterisk) within these sequences. Proximal promoter reflects 1,000 bp. (B) Western blot-based detection of CREB1 and densitometric quantification after transfection of esiRNAs specific to CREB1 in MZ2862RC cell line. The number in parentheses indicates a technical replicate. Same blot was re-probed for β-Actin which served as loading control. Uncropped blot is shown in Supplementary Figure 6. (C) Effect of esiRNA mediated CREB1 knock-down on the expression level of indicated miRNAs. miR-9-5p was used as positive control (n = 3, student t-test). (D) Expression of the indicated miRNAs was determined in 18 CREB1 negative and 18 CREB1 high ccRCC lesions by qRT-PCR and results are visualized as Box-Whisker-Plots (Holm-Sidak method, miR-22: p = 0.269; *p < 0.05).