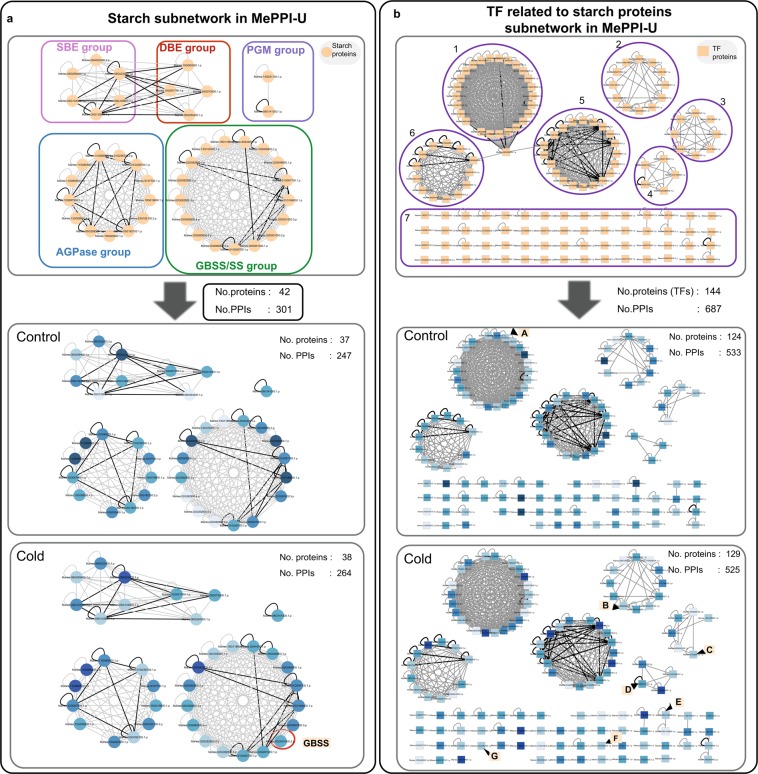
Figure 7.
Dynamic starch and TF subnetworks in cold stressed condition. (a) starch subnetwork was classified into five functional groups; (1) phosphoglucomutase (PGM), (2) glucose-1-phosphate adenylyltransferase (AGPase), (3) starch synthase (SS), (4) 1,4 –alpha-glucan branching enzyme (SBE) and (5) 1,4 –alpha-glucan debranching enzyme (DBE). The red circle represents Manes.02G001000.2.p (GBSS2) protein that showed different interactions relative to control. (b) TFs related to the starch protein subnetwork were separated in 7 groups; (1) ethylene response factor proteins (ERF), (2) Myb domain proteins (MYB), (3) basic helix-loop-helix (bHLH) DNA-binding proteins, (4) NAC domain containing proteins, (5) basic-leucine zipper (bZIP) transcription factor proteins (6) homeobox proteins and (7) other TF proteins. The nodes represent starch (circle) and transcription factor (square) proteins, while the edges show the interactions. The node colors show percentile expression of proteins from highest (dark blue) to lowest (light blue), while the edge colors represent PPIs from both interolog and DDI based predictions (black), or either (grey). The alphabet in orange boxes denote the following TFs proteins; (A) ERF transcription factor protein (ERF; Manes.16G034200.1.p), (B) Myb domain protein 30 (Myb30;Manes.02G046100.1.p), (C) bHLH DNA binding protein (bHLH; Manes.01G269700.1.p), (D) NAC containing protein (NAC;Manes.06G015000.1.p), (E) NIN-like protein 5 (NIN5; Manes.05G130800.2.p), (F) OBF binding protein 4 (OBF4; Manes.06G080600.1.p) and Myb domain protein 96 (Myb96; Manes.06G092600.1.p).