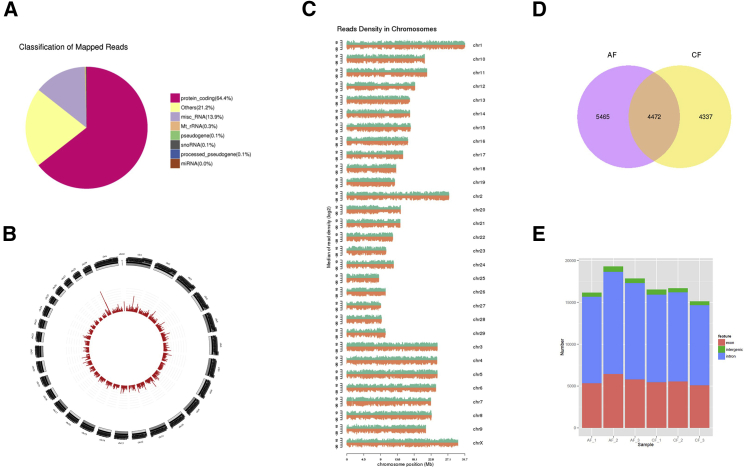
Figure 1.
Identification of circRNAs in Bovine Fatty Tissue
(A) The distribution of reads on the known gene types was obtained according to the expression distribution of each type of gene in the expression quantity statistics samples using HT-Seq software. (B) The total mapped reads were mapped to each chromosome. (C) The total mapped reads were mapped to the density of each chromosome on the genome. We use the ASCII code character sequence for chromosome sorting. (D) Venn diagram depicting different circRNAs uncovered at calf (CF) and adult cattle (AF) subcutaneous fatty tissue. 14,274 circRNA candidates were identified, with 4,337 and 5,465 specific to the calf and adult libraries, respectively. (E) Origin of circRNAs described in this study in the bovine genome.