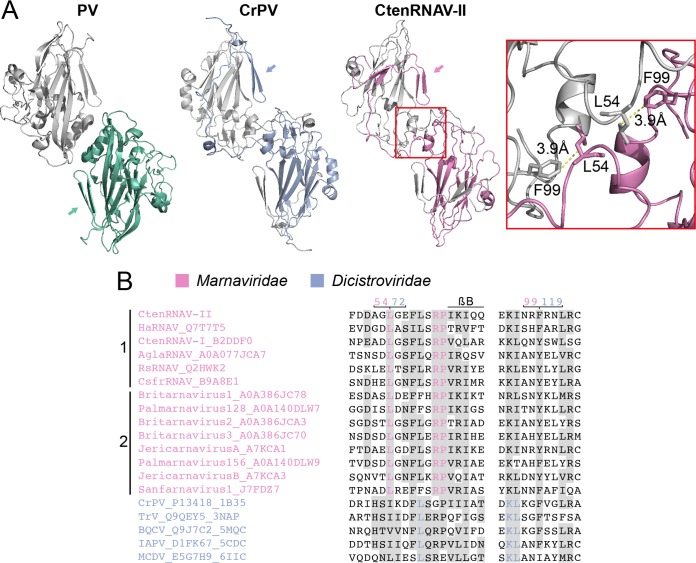
FIG 7.
VP2 N-terminal domain swap. (A) Comparison of two-fold symmetry-related VP2 proteins from poliovirus (PV) (1HXS), Cricket paralysis virus (CrPV) (1B35), and CtenRNAV-II (6SHL) as seen from the inside of the viruses. The arrows indicate the location of the N termini of the bottom VP2 proteins of each virus, color-coded green, blue, and pink, respectively. PV does not contain the domain swap, but CrPV and CtenRNAV-II do. The two residues, L54 and F99, of CtenRNAV-II that might be of importance for the domain swap are represented as sticks in the right-most image. The distance between L54 from one of the VP2 proteins to F99 of the other VP2 protein is 3.9 Å. (B) Selected areas from a sequence alignment (76) of VPs from representatives of viruses that are currently classified by ICTV to the Marnaviridae family and viruses of the Dicistroviridae family that have structures available in the PDB. The indication of a secondary structure above the alignments follows CtenRNAV-II and is the same as in PDB entry 6SHL. Six of the Marnaviridae viruses, including CtenRNAV-II, are identified as diatom viruses (1), whereas the other eight were discovered through metagenomic analysis (2). At least 75% identities, either within each virus family or within the whole alignment, are colored in pink and blue, respectively, and similarity (80) across each family or across the entire alignment is shaded gray. The residue numbering follows CtenRNAV-II (pink) and CrPV (blue).