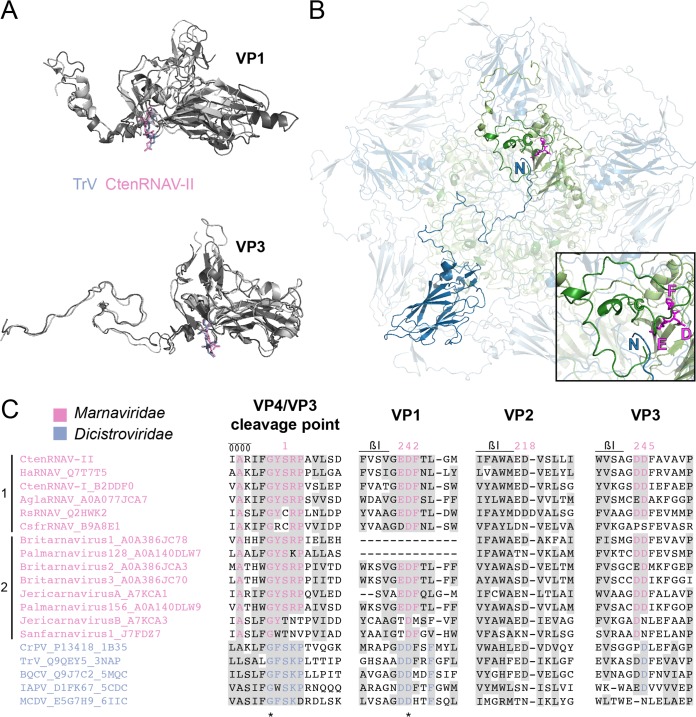
FIG 8.
Putative autoproteolytic motifs in VP1 and VP3. (A) Superimposition of VP1 and VP3 proteins of CtenRNAV-II (dark gray) and Triatoma virus (TrV) (3NAP) (light gray). The catalytic motifs are shown in stick representation in pink and blue, respectively. (B) A CtenRNAV-II pentamer viewed from the inside of the capsid. The C terminus of a VP4 protein (dark green) and N terminus of a VP3 protein (dark blue), located in an adjacent five-fold related protomer, are marked. The position of the catalytic motif EDF in VP1 (light green) is highlighted in magenta, where the glutamic acid points toward the gap between VP4 and VP3. The inset shows a closeup of VP4, the N terminus of VP3, and the EDF motif in VP1. (C) Selected areas from a sequence alignment (76) of VPs from representatives of viruses that are currently classified by ICTV to the Marnaviridae family and viruses of the Dicistroviridae family that have structures available in the PDB. The indication of a secondary structure above the alignments follows CtenRNAV-II and is the same as in PDB entry 6SHL. Six of the Marnaviridae viruses, including CtenRNAV-II, are identified as diatom viruses (1), whereas the other eight were discovered through metagenomic analysis (2). At least 75% identity, either within each virus family or within the whole alignment, is colored in pink and blue, respectively, and similarity (80) across each family or across the entire alignment is shaded gray. Identical residues among all sequences are marked with an asterisk. The residue numbering follows CtenRNAV-II.