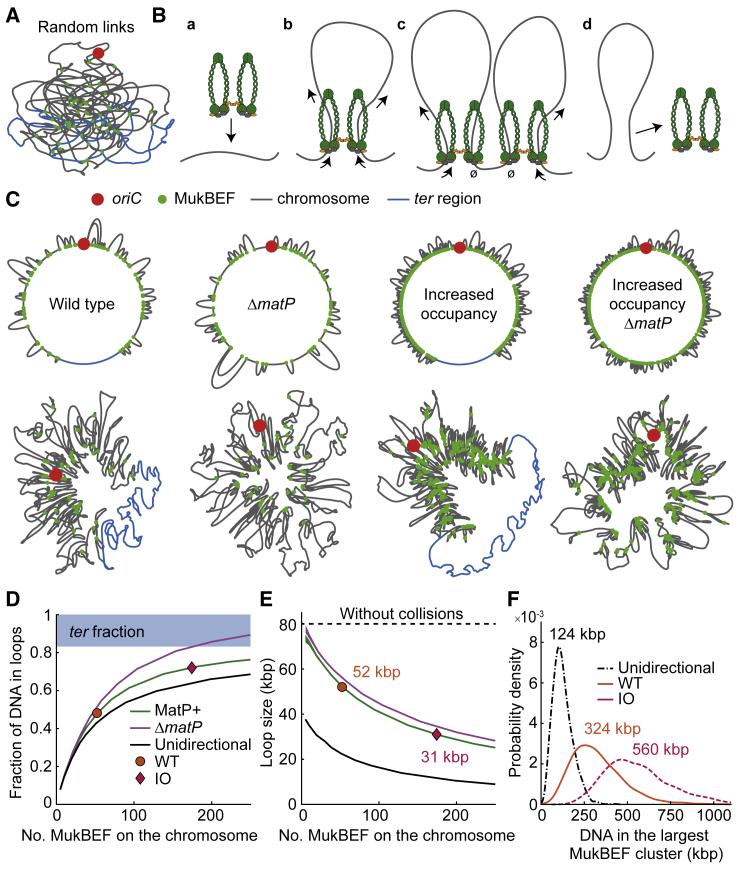
Figure 5.
Modeling Loop Extrusion by MukBEF
(A) Example of a simulated randomly linked E. coli chromosome.
(B) Description of the model: (a) A MukBEF dimer of dimers associates at a random site on the chromosome. (b) The loop is enlarged by the two dimers moving in opposite directions along the chromosome. (c) Collisions between MukBEFs prevent loop extrusion only on the internal collided dimers. (d) MukBEF spontaneously dissociates, releasing the loop. The MatP-bound ter region immediately displaces MukBEF upon contact.
(C) Representative E. coli chromosomes with and without MukBEF displacement from ter and WT or increased MukBEF occupancy. (top) Beginning and end of loops with MukBEF (green dots) along the chromosome. (bottom) Force-directed layouts of the chromosomes.
(D and E) (D) Fraction of the chromosome within a loop and (E) loop size per individual MukBEF as a function of the number of chromosome-bound MukBEF dimers of dimers. In unidirectional loop extrusion, dimers are not connected and each dimer acts independently, binding and extruding a loop in a randomly chosen direction. Estimated numbers for MukBEF for WT and IO cells are shown. Line thickness denotes 95% bootstrap confidence interval for the mean across 1,000 simulation replicas.
(F) Distribution of DNA in the largest MukBEF cluster (no unlooped DNA between them) for WT and IO occupancy and unidirectional loop extrusion (from the same data as in E).