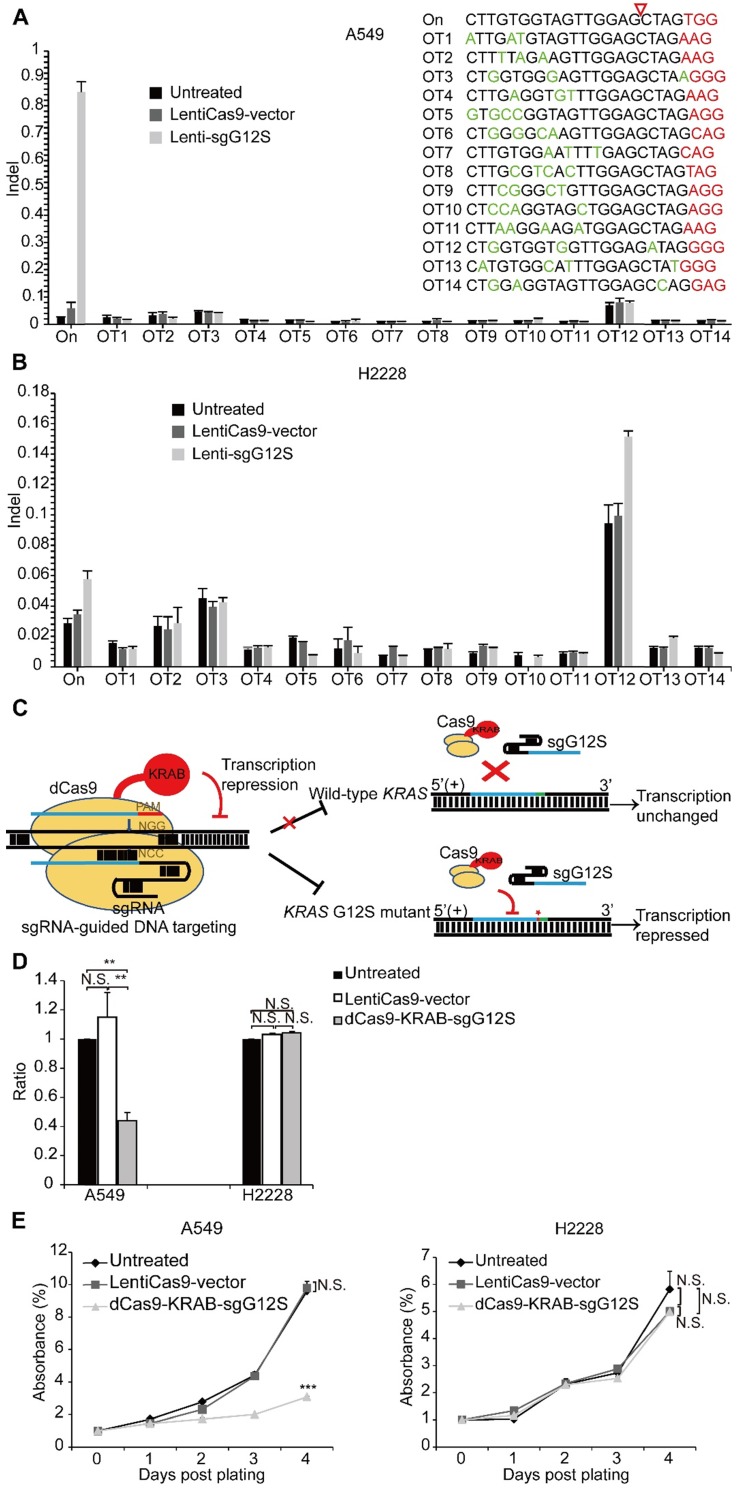
Figure 3.
dCas9-KRAB mRNA-regulating system downregulated G12S transcription and inhibited tumor cell proliferation. (A, B) No off-target indels were noticeably caused by the CRISPR/Cas9 gene-cutting system at fourteen homologous sites that differed from the on-target sites by up to 4 nt. PAM sequences are shown in red and mismatched nucleotides are shown in green. On: on-target site. OT: off-target site. Cleavage position within the 20 bp target sequences is indicated by a red arrow. Error bar indicates S.E.M. (n=3 to 4). (C) Diagram of knocking down KRAS G12S mutant allele specifically by the dCas9-KRAB system. Blue strands: spacer; green strands: PAM sequence; red strands and star: single-nucleotide missense mutations. (D) qRT-PCR analysis of KRAS G12S mRNA expression. Error bars represent S.E.M. (∗) 0.01<P < 0.05, (∗∗) 0.001<P < 0.01, (∗∗∗) P < 0.001. (E) CCK-8 assay. Cell proliferation was determined at different timepoints by CCK-8 reagents. The relative number of cells of each group with different treatments was determined by normalizing the optical density at 490 nm of each CCK-8 reaction to the average optical density of the negative control groups.