Figure 5. Phylogenetic relationship of Mxra8 orthologs.
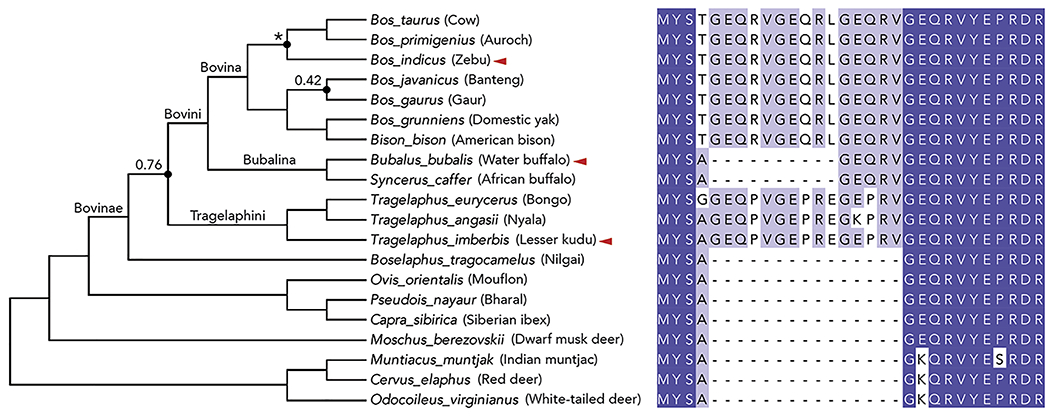
The phylogeny summarizes the relationships for Bovinae, Bovidae, Moschidae, and Cervidae as estimated by (Zurano et al., 2019) but was modified to include the relationships for Bos taurus and Bos indicus (reported by (Wang et al., 2018)) as indicated by asterisks. All nodes have high support with posterior probabilities >95% unless indicated otherwise by values. Species indicated with red arrows are investigated in subsequent functional experiments. The phylogenetic tree and sequence alignment were prepared using FigTree and Jalview (Waterhouse et al., 2009), respectively. Protein sequences of Mxra8 gene orthologs from Bovidae members in the region corresponding to the 15-residue Mxra8 insertion in D1 were aligned using MUSCLE and visualized using Jalview. Amino acid residues are colored according to sequence identity among Bovidae family members: dark blue boxes (100%), light blue boxes (50-90%), and white boxes (<50%). Gaps in the protein sequences are indicated by a dash.
