Table 1.
Bayesian selection of the relaxed-clock model
| Data | Model | log mL 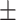 S.E S.E |
Pr |
|---|---|---|---|
| 1g, 20s | AR |
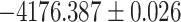
|
0.993 |
| IR |
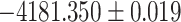
|
0.007 | |
| STR |
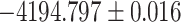
|
0 | |
| 1g, 40s | AR |
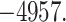 026 026 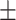 0.050 0.050 |
1 |
| IR |
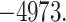 258 258 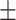 0.040 0.040 |
0 | |
| STR |
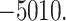 601 601 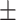 0.047 0.047 |
0 | |
| 1g, 85s | AR |
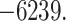 864 864 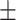 0.059 0.059 |
1 |
| IR |
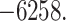 492 492 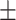 0.069 0.069 |
0 | |
| STR |
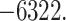 348 348 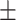 0.043 0.043 |
0 | |
| 5g, 20s | AR |
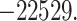 810 810 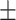 0.035 0.035 |
0.999 |
| IR |
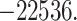 580 580 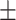 0.030 0.030 |
0.001 | |
| STR |
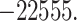 840 840 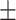 0.022 0.022 |
0 | |
| 20g, 20s | AR |
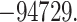 470 470 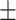 0.043 0.043 |
0.998 |
| IR |
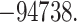 010 010 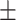 0.058 0.058 |
0.002 | |
| STR |
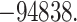 540 540 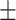 0.038 0.038 |
0 | |
| 20g, 40s | AR |
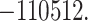 300 300 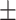 0.181 0.181 |
1 |
| IR |
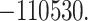 800 800 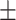 0.218 0.218 |
0 | |
| STR |
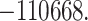 500 500 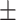 0.130 0.130 |
0 |
Notes: Data list each treatment with the number of genes (g) and species (s) for each alignment. Models tested include AR, autocorrelated rates; IR, independent rates; STR, strict clock. Log mL + SE is the log-marginal likelihood for the model with standard error for the log-likelihood estimate. Pr is the posterior model probability (assuming equal prior probabilities for models), calculated as in dos Reis et al. (2018, Appendix 2).
