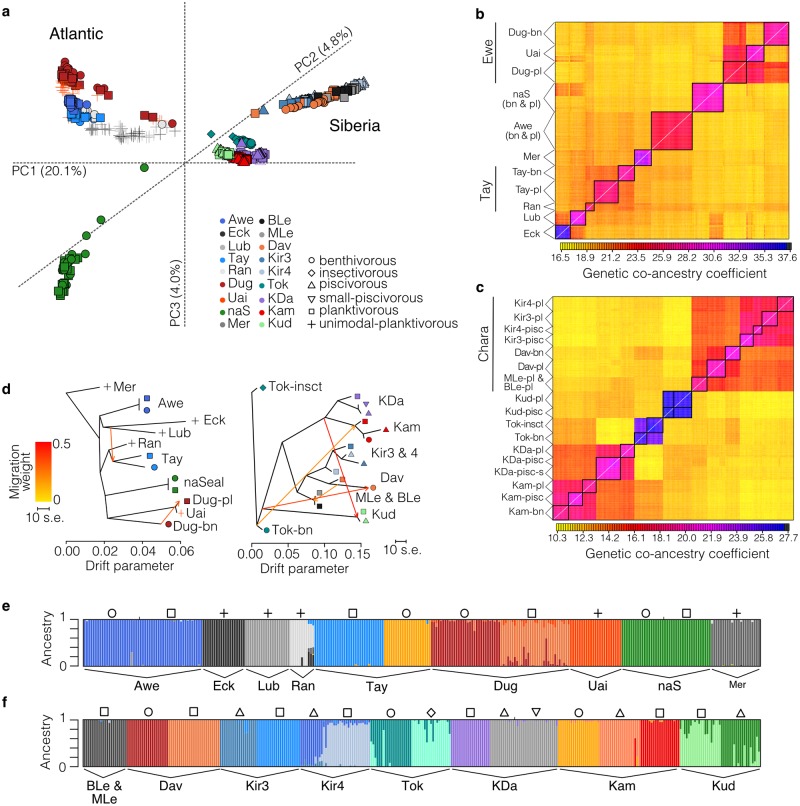
Fig 3. Hierarchical population genetic structure and genomic background divergence.
(A) 3-dimensional principal components plot for all individuals (N = 630) based on 12,215 SNPs. Lake abbreviations are explained in S1 Table. PC1 mostly separates the Atlantic and Siberian lineage, PC2 mostly separates catchments within the Siberian lineage and PC3 separates na Sealga from all other populations within the Atlantic lineage (B,C) fineRADstructure results showing co-ancestry coefficients between individuals from the (B) Atlantic and (C) Siberian lineage. Ecotypes or lake populations that form discrete genetic clusters are enclosed in black boxes. Note the high genetic co-ancestry across lakes within the same catchment, such as (B) Dughaill (Dug) and Uaine (Uai) in the Ewe catchment or (C) Kiryalta-3 (Kir3), Kiryalta-4 (Kir4), Davatchan (Dav), and Maloe and Bol’shoe Leprindo (MLe, BLe) in the Chara catchment. Ecotype descriptions (abbreviations in S1 Table) are shown unless populations were unimodal or ecotype-associations unknown. Populations are ordered in the same way along the x-axis of the co-ancestry matrices. (D) Allele frequency-based maximum-likelihood trees showing the most likely phylogenetic relationships across lakes and ecotype pairs by lineage, including two and four fitted migration events in the Atlantic and Siberian lineage, respectively. Migration events are shown as arrows coloured by migration weight. (E,F) Admixture plots showing the genetic ancestry for all individuals from the (E) Atlantic and (F) Siberian lineage for K = 11 and K = 16, respectively. Ecotypes are marked by symbols above each cluster.