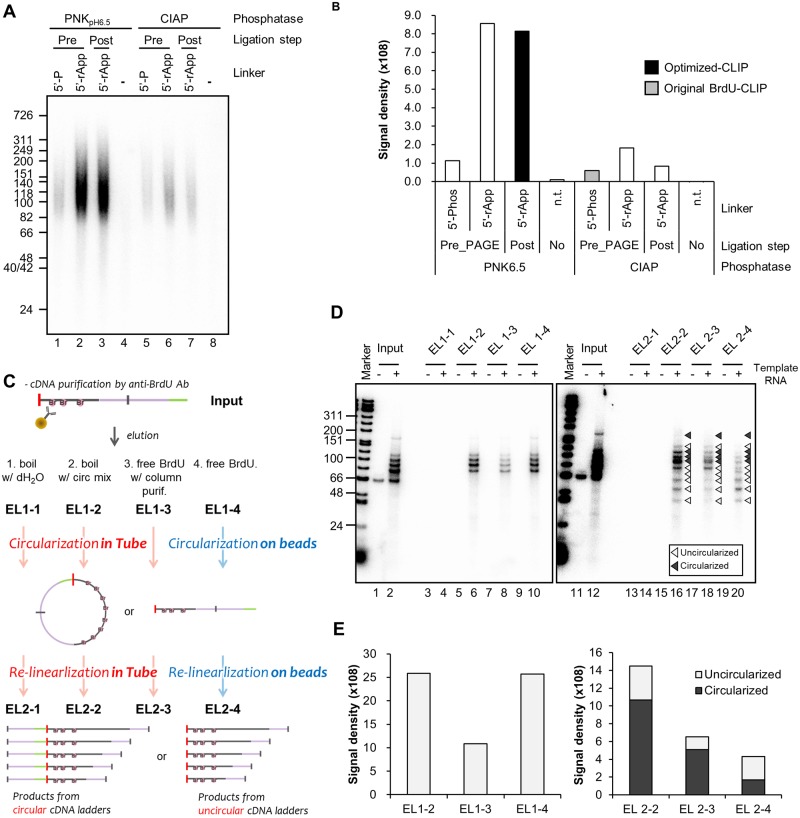
Fig 2. Optimization of dephosphorylation, linker ligation and circularization.
(A–B) Quantification of reverse transcribed cDNA to evaluate the yield of linker-ligated RNA. Template RNA was isolated from HNRNPU-RNA, dephosphorylated with PNK or CIAP, and subsequently ligated using linkers with phosphorylated or adenylated 5’-ends. Linkers were phosphorylated or adenylated prior to polyacrylmide gel electrophoresis (PAGE) or after isolation from the membrane. cDNA that was reverse transcribed in the presence of [α-32P]-dCTP was purified with anti-BrdU beads and separated in a 10% TBE-urea gel (A). Signal density was quantified from the autoradiograph (B). (C) Schematics for optimization of elution during anti-BrdU purification and during circularization using RNA ladders. (D–E) Reverse transcribed cDNAs from linker-ligated RNA ladders were eluted from anti-BrdU beads as indicated in (C), and were subsequently circularized and re-linearized in tubes (EL2-1, -2, -3) or on beads (EL2-4).