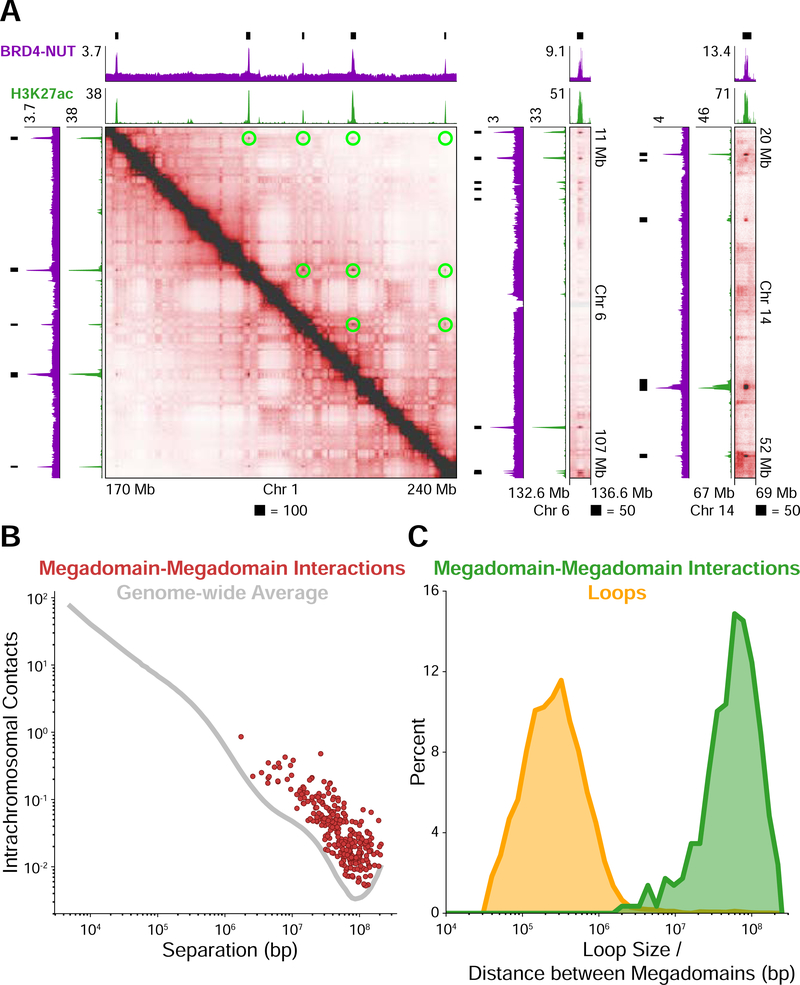
Figure 1. Distant Interactions Between Broad, Hyperacetylated Chromatin Domains.
(A) Hi-C contact maps at 100 kb resolution for the patient-derived TC-797 cell line endogenously expressing BRD4-NUT. BRD4-NUT (magenta) and histone H3K27ac (green) ChIP-seq profiles are aligned with the maps. Black bars indicate megadomains. Megadomain-megadomain interactions are indicated by green circles above the diagonal in the leftmost map; below the diagonal is unannotated for clarity. The rightmost two maps are additional examples displayed as off-diagonal slices of the Hi-C contact map.
(B) Intrachromosomal contacts for loci separated by the distance given on the abscissa. The genome-wide average is indicated as a gray line. Each megadomain-megadomain interaction is displayed as a red dot indicating that interactions between megadomains are stronger than the genome-wide average for loci separated by the same distance.
(C) Percent of megadomain-megadomain interactions (green) separated by the distance given on the abscissa and percent of chromatin loops (yellow) of a given size. Distances between interacting megadomains are orders of magnitude larger than chromatin loops. Sizes/distances are logarithmically binned.