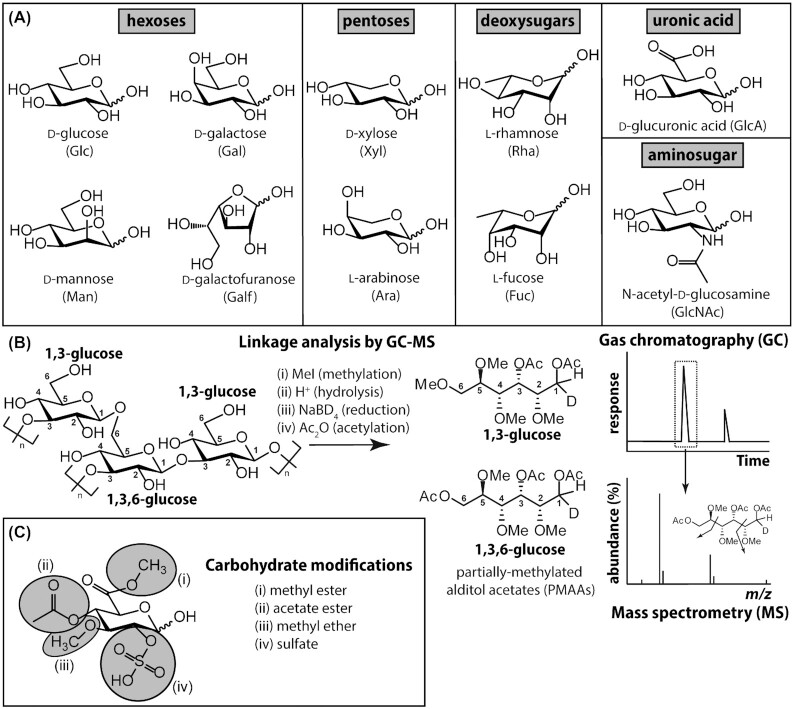
Figure 3.
Common monosaccharide building blocks of lichen polysaccharides. (A) Although hundreds of different monosaccharides have been found in nature, only a fraction of them have been identified in lichen polysaccharides. These monosaccharides may be classified as neutral hexoses or pentoses (six and five carbon sugars, respectively), deoxyhexoses [in which the carbon 6 (C6) hydroxy group is removed], hexosamines or aminosugars (bearing an acetylated amino group at C2) and uronic acids (in which C6 bears a carboxylate moiety). Note that for simplicity, throughout the text the absolute configurations (D or L) are omitted from the full monosaccharide name or abbreviation; likewise, a pyranose (i.e. six-membered) ring configuration is assumed for all monosaccharides with the exception of galactofuranose (Galf). Refer to Varki et al. (2015, 2017) for common monosaccharide and polysaccharide conventions. (B) The monosaccharide constituents of lichen polysaccharides, and details concerning the regiochemisty of how they are glycosidically linked, may be deduced using GC-MS-based linkage (or methylation) analysis, shown here for a hypothetical β-glucan fragment. In brief, unsubstituted hydroxy groups are methylated (i) prior to complete polysaccharide hydrolysis under strongly acidic conditions (ii). Subsequent reduction using sodium borodeuteride (iii) tags the reducing end of each monosaccharide and acetylation with acetic anhydride (iv) caps hydroxyl groups that had been protected from methylation by virtue of the fact that they were glycosidically linked. The partially methylated alditol acetates (PMAAs) so created are resolvable by GC and may be identified, using appropriate standards, based on their predictable fragmentation by MS. Note that this technique is unable to assign anomeric (i.e. α or β) configurations. (C) Many lichen-derived monosaccharides bear biochemically modified hydroxy groups, represented here for a fictitious glucuronic acid. Most of these modifications (i, ii and iv) are acid labile and undetected by linkage analysis, unless substantially modified protocols are used.