Figure 1.
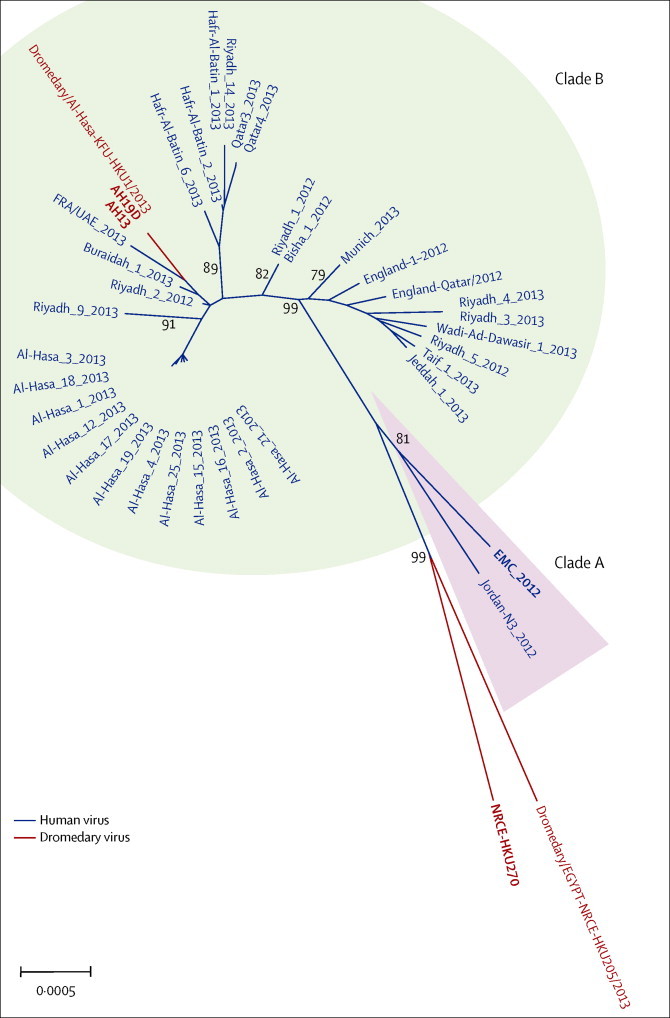
Phylogenetic tree of MERS-CoV full genome sequences
Virus isolates used for this study are in bold. The tree was built by the maximum likelihood method in Mega 6.06 with 500 bootstrap replications. Bootstrap values are shown at the major nodes of the tree. The scale bar shows the mutation rate for the tree. Accession numbers of virus genomes retrieved for this analysis are JX869059, KC776174, KC164505, KC667074, KF192507, KJ156881, KJ156949, KJ156944, KJ556336, KJ156952, KF600613, KF186564, KF600627, KF186567, KF600651, KJ156866, KF600634, KF600632, KF600644, KF186565, KF186566, KF600645, KF600647, KF600630, KF600652, KF745068, KJ156869, KJ650297, KJ650296, KJ650295, KF600628, KF961221, KF961222, KJ156874, KJ156910, KJ156934, KF600612, KF600620, and KJ477102. MERS-CoV=Middle East respiratory syndrome coronavirus. EMC=human MERS-CoV EMC. AH13=MERS-CoV Dromedary/Al-Hasa-KFU-HKU13/2013. AH19D=MERS-CoV Dromedary/Al-Hasa-KFU-HKU19D/2013. NRCE-HKU270=MERS-CoV Dromedary/Egypt-NRCE-HKU270/2013.
