Abstract
Introduction
Low gut microbiome richness is associated with dyslipidemia and insulin resistance, and ceramides and other sphingolipids are implicated in the development of diabetes.
Objectives
Determine whether circulating sphingolipids, particularly ceramides, are related to alterations in the gut microbiome among obese patients with increased diabetes risk.
Methods
This was a cross-sectional and longitudinal retrospective analysis of a dietary/weight loss intervention. Fasted serum was collected from 49 participants (41 women) and analyzed by HPLC-MS/MS to quantify 45 sphingolipids. Shotgun metagenomic sequencing of stool was performed to profile the gut microbiome.
Results
Confirming the link to deteriorated glucose homeostasis, serum ceramides were positively correlated with fasting glucose, but inversely correlated with fasting and OGTT-derived measures of insulin sensitivity and β-cell function. Significant associations with gut dysbiosis were demonstrated, with SM and ceramides being inversely correlated with gene richness. Ceramides with fatty acid chain lengths of 20–24 carbons were the most associated with low richness. Diet-induced weight loss, which improved gene richness, decreased most sphingolipids. Thirty-one MGS, mostly corresponding to unidentified bacteria species, were inversely correlated with ceramides, including a number of Bifidobacterium and Methanobrevibacter smithii. Higher ceramide levels were also associated with increased metagenomic modules for lipopolysaccharide synthesis and flagellan synthesis, two pathogen-associated molecular patterns, and decreased enrichment of genes involved in methanogenesis and bile acid metabolism.
Conclusion
These data establish a link between gut microbiota richness, ceramides, and diabetes risk in overweight/obese humans, and implicate the gut microbiota in the dysregulation of lipid metabolism in metabolic disorders.
Keywords: Microbiome, ceramides, sphingolipids, endotoxin, glucose metabolism
1. Introduction
Type 2 diabetes is the result of insulin resistance and deterioration of β-cell function. Yet the environmental factors, and their links to physiological mediators, that drive the development of type 2 diabetes are only partially understood. The gut microbiota has emerged as a potential contributor to the regulation of glucose homeostasis. Type 2 diabetes is associated with alterations in a number of bacteria and their biological functions in both Chinese (Qin et al. 2012) and European (Karlsson et al. 2013) populations. Metabolic changes that precede frank diabetes can also be transferred via the gut microbiota to mice (Ridaura et al. 2013). Low gut microbiota richness, which is altered in some moderate obesity/overweight patients without overt diabetes (Cotillard et al. 2013), may be an important factor in the progression to diabetes.
The microbial ecosystem can be characterized by its richness i.e. the number of unique bacterial genes present in a metagenome. People in Western societies, with high rates of obesity, type 2 diabetes, and frequent exposure to antibiotics and food additives that can impact gut microbes, have considerably lower bacterial diversity than previously unexposed hunter-gatherers (Clemente et al. 2015). Gastrointestinal diseases such as liver cirrhosis (Qin et al. 2014) and inflammatory bowel diseases (Qin et al. 2010) have been associated with a loss of metagenomic richness. Metabolic characteristics are also altered in individuals with low gut microbiome richness, as Danish subjects with fewer microbial genes have increased BMI, insulin resistance, inflammatory markers, as well as hypertriglyceridemia (Le Chatelier et al. 2013). These associations were also observed in French individuals and our laboratory further showed that this was partially reverses by diet-induced weight loss (Cotillard et al. 2013). Lipid metabolism is critical to the pathogenesis of diabetes (Glass & Olefsky 2012) and alterations in the gut microbiota could potentially regulate these pathways.
Sphingolipids are a diverse class of membrane lipids that also act as intracellular signaling molecules. Ceramides, which accumulate with lipid loading and inflammation, are increased in patients with type 2 diabetes (Haus et al. 2009; Shui et al. 2013), and they can directly induce insulin resistance (Stratford et al. 2004) and regulate β-cell apoptosis (Lupi et al. 2002). Plasma ceramides may even become clinical biomarkers, but the environmental factors that contribute to ceramide accumulation, outside of obesity, are not well defined (Summers 2018). It was recently shown in mice, however, that the gut microbiota regulates liver ceramide synthesis and subsequent hepatic steatosis and gluconeogenesis through FXR-dependent secondary bile acid metabolism (Jiang et al. 2015). Furthermore, endotoxin-induced inflammation, a potentially important link between impaired glucose homeostasis and gut dysbiosis (Cani et al. 2007a), readily stimulates sphingolipid synthesis. It is still unknown, however, whether an altered gut microbiota is associated with increased ceramide levels in humans. Combining targeted lipidomics and quantitative metagenomics, the objective of the present study was to examine whether ceramide accumulation could link gut dysbiosis to increased diabetes risk in individuals with overweight or obesity.
2. Methods
2.1. Study population
The study was conducted at Pitié-Salpêtrière Hospital (Nutrition department) in Paris, France. Forty-nine participants were overweight (N=11) and obese (N=38) (8 males, 41 females), and have been described in detail elsewhere (Cotillard et al. 2013; Kong et al. 2014). Participants did not have diabetes, chronic or inflammatory diseases, or related treatments, and did not take antibiotics within the two months prior to stool collection.
2.2. Dietary intervention
The intervention consisted of a 6-week period on a low calorie diet enriched with soluble fibers, high protein intake, and low glycemic index carbohydrates. Daily caloric intake was approximately 1,200 kcal/d for women and 1,500 kcal/d for men, composed of 35% protein, 25% fat and 40% carbohydrate. A 6-week weight stabilization diet followed the calorie restriction period, and was established based on each subject’s resting energy expenditure and physical activity levels.
2.3. Body composition, adipose tissue, and biochemical parameters
Details of the analyses have been described elsewhere (Rizkalla et al. 2012; Cotillard et al. 2013). Body composition was determined using dual energy x-ray absorptiometry for the estimation of total body fat and android fat proportion. Subcutaneous adipose tissue samples were evaluated for adipocyte size and immune cell frequency (Aron-Wisnewsky et al. 2009). Blood samples were collected after an overnight fast to measure routine biochemical parameters. Serum insulin was assayed by a chemiluminescense (ARCHITECT system, Abbott).
Insulin sensitivity was quantified using the following fasting indices: the Homeostasis Model Assessment of insulin sensitivity (HOMA-S), calculated using the HOMA2Calculator system (Wallace et al. 2004); Quantitative Insulin Sensitivity Check Index (QUICKI) (Katz et al. 2000); and the Disse index (Antuna-Puente et al. 2009), a lipid-based index of insulin sensitivity
An oral glucose tolerance test (OGTT) was performed in 40 subjects (39 that had insulin measured during the test at all time points). The Matsuda Index was calculated as a measure of insulin sensitivity (Matsuda & DeFronzo 1999). β-cell function was estimated by the insulin secretion and sensitivity index-2 (ISSI-2), an OGTT-analog of the IVGTT-derived disposition index. It is the product of insulin secretion in response to glucose, measured as the ratio of insulin AUC to the glucose AUC, and the Matsuda index. The ISSI-2 captures the hyperbolic relationship between insulin sensitivity and secretion (Retnakaran et al. 2008) and correlates with the disposition index (Retnakaran et al. 2009). We confirmed the hyperbolic relationship in our sample population, as outlined in the original paper (data not shown) (Retnakaran et al. 2008).
2.4. Targeted lipidomics
Analysis of sphingolipids was conducted by HPLC-MS/MS, as described previously (Camont et al. 2013), in overnight fasted serum. Lipids were extracted in acidified methanol:chloroform with both internal and calibration standards (Avanti Polar Lipids, Alabaster, AL, USA). Forty-five sphingolipids were quantified, namely sphingomyelins (SM), dihydroceramides (DHCer), defined as Cer(d18:0) in standard nomenclature, and sphingosine- or sphingadienine-containing ceramides, that is Cer(d18:1) and Cer(d18:2), respectively. These 45 lipids were detected in at least 80% of samples. To avoid bias introduced by simple 0-imputation of the values below detection, measurements below the level of quantitation were imputed using multiplicative log-normal-randomized imputation with the zCompositions package in R.
2.5. Faecal metagenomic analysis
Total faecal DNA was extracted and sequenced on a high-throughput ABI SOLiD sequencer. Reads were cleaned for quality and origin (human, cow, and plant), mapped to the 3.9 million gene reference catalogue (Nielsen et al. 2014) and normalized by nucleotide length. Forty-five subjects with sufficient sampling depth were used for subsequent analyses. Gene richness was calculated, after downsizing to 7 million genes, as the total number of uniquely mapped genes per sample. Richness categories were split at 460,000 genes in the 3.3M catalog, as done previously (Cotillard et al. 2013). Metagenomic species (MGS) were constructed from total-sum normalized gene abundance data using the catalog provided by Nielsen et al. and only co-abundance clusters greater than 700 genes were considered as bacterial species (Nielsen et al. 2014). Computations were performed using the [momr] R package (Le Chatelier et al. 2013; Prifti et al. 2013)
Kegg Ortholog (KO) annotation was provided by the reference catalog, and each gene was summed into its respective KO. Annotation of bile acid genes was performed as described by others (Tremaroli et al. 2015). Briefly, protein sequences from organisms containing complete bile acid pathways were extracted, specifically: Clostridium scindens ATCC 35704, Bifidobacterium longum SBT2928, Bacteroides fragilis NCTC9343, Eggerthella lenta DSM 2243, and Ruminococcus gnavus ATCC 29149. Sequences were then aligned to the reference catalog using UBLAST with an E-value cutoff of 10-5. These candidates were then aligned against the KEGG database to include only genes with best hits to bile acid proteins. Bile acid genes were grouped by their functional annotation in MetaCyc (e.g. bile acid degradation).
2.6. Statistical Analysis
Statistical analyses were performed in the R Language and Environment with packages listed in [brackets]. As the data were generally positively skewed (lipidomics) or non-normal (metagenomics), robust statistics were used when possible. The Spearman method was used for correlations. The MM-type robust regression [lmrob function in robustbase] was used for multiple regression (Yohai 1987). Longitudinal data were analyzed using mixed effects models with random intercepts for subject [lme4, car, multcomp]. Enrichment of KEGG modules and pathways were tested with the reporter features algorithm using adjusted p-values and coefficients from the correlations of each KO to Cer(d18:1) [piano] (Karlsson et al. 2013). Only distinctly up- and down-regulated gene sets are reported. The null distribution was used for the significant assessment of gene sets between 3 and 100 KOs. For clinical (including gene richness) and lipid class associations, unadjusted p-values were reported. However, given the a priori nature of metagenomic analysis, p-values for tests of MGS or KOs was adjusted for the Benjamini-Hochberg false discovery rate; the only exception being bile acid gene set enrichment, as this hypothesis was pre-specified (Jiang et al. 2015).
3. Results
3.1. Circulating ceramides associate with a host of metabolic alterations in persons with overweight/obesity
Characteristics of the population used for lipidomics profiling have been described previously (Cotillard et al. 2013), and are summarized in Table 1 and Online Resources 1 and 2. Median BMI was approximately 33 kg/m2 and body fat was 40%. None of the subjects had diabetes. Shotgun metagenomic analysis indicated that 41 women had a median gene richness of 507,800 and 8 men had a median gene richness of 537,000, which were not statistically significantly different. We confirmed that gene richness was inversely associated with fasting TG (r=−0.46, P<0.001), as previously reported.
Table 1.
Clinical characteristics of the study population and correlations to gene richness.
| Summary statistics | Correlation to gene richness | ||||
|---|---|---|---|---|---|
| Variable (units) | 1Q | Median | 3Q | Rho | P |
| Age (years) | 32 | 41 | 55 | 0.01 | 0.956 |
| Weight (kg) | 79.7 | 93.1 | 101 | −0.06 | 0.688 |
| BMI (kg/m2) | 30.49 | 33.2 | 36.6 | −0.11 | 0.458 |
| Body fat (%) | 36.8 | 40.04 | 43.76 | −0.02 | 0.9 |
| Total cholesterol (mM) | 4.71 | 5.15 | 5.78 | −0.26 | 0.087 |
| HDL-C (mM) | 1.14 | 1.35 | 1.61 | 0.06 | 0.688 |
| LDL-C (mM) | 2.95 | 3.2 | 3.71 | −0.1 | 0.531 |
| TG (mM) | 0.76 | 1.08 | 1.46 | −0.46 | 0.001 |
| Glucose (mM) | 5 | 5.2 | 5.4 | −0.19 | 0.201 |
| Insulin (mU/L) | 6 | 8.3 | 10.6 | −0.19 | 0.219 |
| CRP (mg/L) | 1.3 | 3.6 | 7.06 | −0.18 | 0.235 |
All 49 subjects had each clinical measurement. Forty-five subjects had sufficient metagenomic sampling depth to calculate gene richness and the subsequent correlation. Rho reflects Spearman correlations.
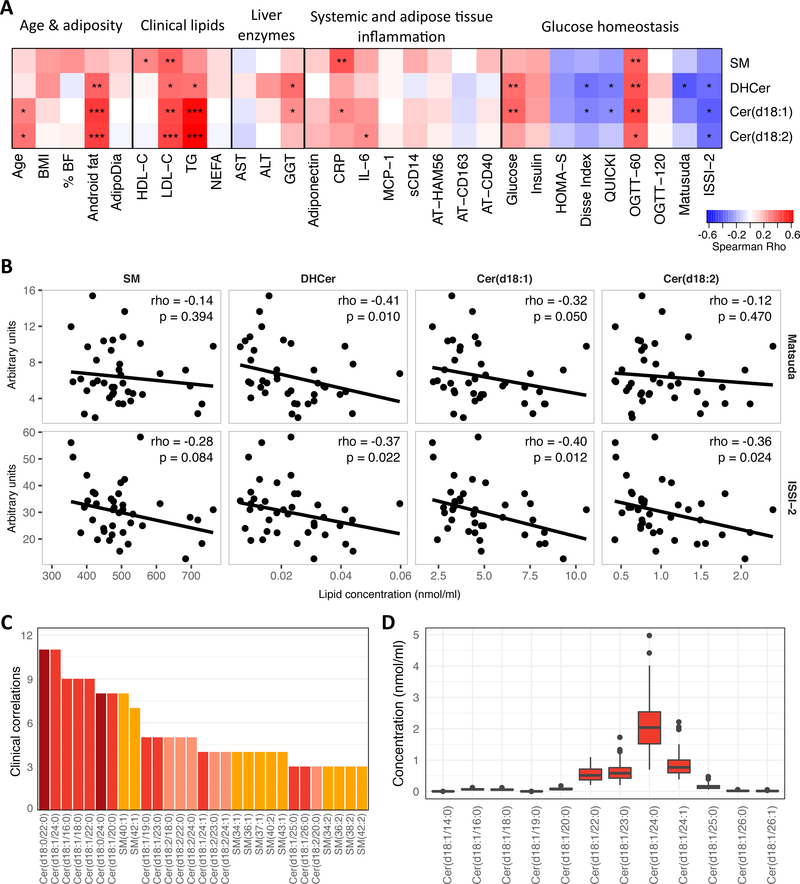
Serum sphingolipids were tested for associations with a number of diabetes risk factors. Four sphingolipid subclasses were quantified, covering a total number of 45 lipids including sphingomyelins (SM), dihydroceramides (DHCer) or Cer(d18:0) in standard nomenclature, and (d18:1)Cer or (d18:2)Cer ceramides, which correspond to a classical sphingosine backbone or a monounsaturated variation, respectively. The most significant associations found were with the non-SM subclasses and clinical variables linked to deteriorated metabolic homeostasis such as accumulation of android fat, age, cholesterol, triglycerides, and GGT (Figure 1A). Importantly, regarding glucose homeostasis, all subclasses were positively associated with 60 min glucose during the OGTT, but only two subclasses, DHCer and Cer(d18:1), were associated with impaired insulin sensitivity determined by both fasting (Disse, QUICKI) and the OGTT-derived Matsuda index (Figure 1B). Furthermore DHCer, Cer(d18:1), and Cer(d18:2), inversely associated with the ISSI-2 index, indicative of impaired β-cell function (Figure 1B). SM, Cer(d18:1) and Cer(d18:2) were also positively correlated with circulating CRP or IL-6.
Figure 1.
A: Spearman correlations between clinical variables and sphingolipid subclasses. *P<0.05, **P<0.01, ***P<0.001. % BF = Percent body fat, AdipoDia = adipocyte diameter, TG = fasting triglycerides, NEFA = non-esterified free fatty acids, sCD14 = soluble CD14, OGTT-60 and OGTT-120 = plasma glucose during OGTT at 60 and 120 minutes, respectively, ISSI-2 = insulin-secretion-sensitivity-index-2. B: Lipid subclasses vs. OGTT-derived Matsuda index (top) and ISSI-2 (bottom). Rho and p-values from spearman correlations. Regression line is a robust regression fit with the R function ‘rlm’. C: Sum of significant associations (P<0.05) for spearman correlation of each lipid species to each non-lipid (e.g. HDL-C) clinical parameter. See Online Resource 3w for complete results. Lipid species are colored by subclass. DHCer are labeled by their chemical structure Cer(d18:0). D: Absolute serum concentration of individual Cer(d18:1) species.
In order evaluate the relative weight of each sphingolipid subclass in the association with clinical parameters, the sum of the number of variables (excluding cholesterol and TG) with significant associations with individual shphingolipid species was calculated. This revealed that DHCer and Cer(d18:1) were more strongly associated with clinical parameters than other species; together they accounted for 7 of the top 9 lipids with the highest number of non-lipid (e.g. HDL-C) clinical correlations (Figure 1C; Online Resource 3). Importantly, very-long chain ceramides (C22 – C24 species), which are the most abundant species in human serum (Figure 1D), were as related to clinical phenotype as C16 and C18 species. Overall, these data confirm that serum ceramides levels are associated to a deleterious metabolic profile.
3.2. Serum ceramides are negatively correlated to gut microbiota richness
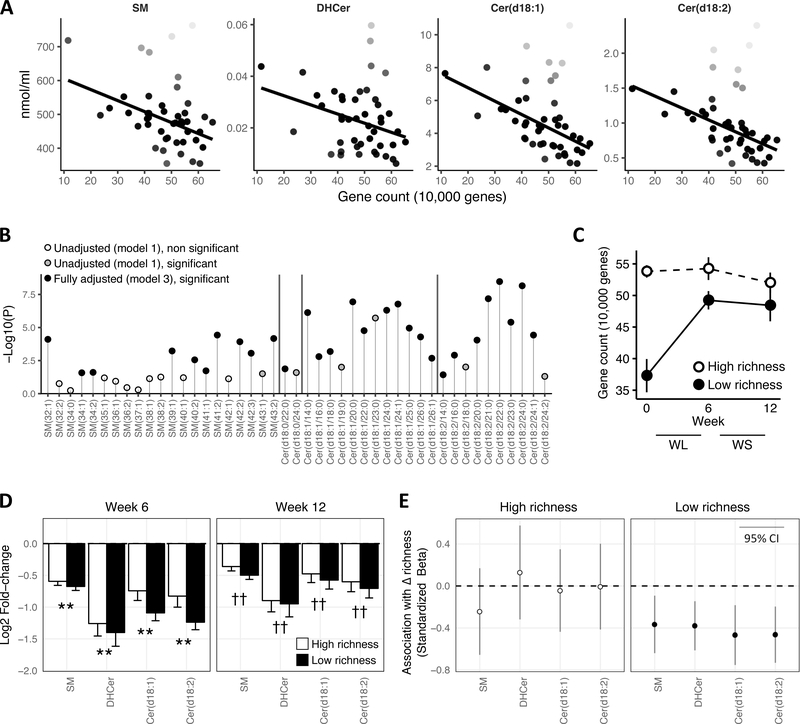
Sphingolipids may be an important component of the dyslipidemia associated with low bacterial richness. Consistent with this hypothesis, there was an inverse association between gene richness and each lipid subclass, although the statistical significance was strongest for Cer(d18:1) (rho=−0.45, P=0.002) and Cer(d18:2) (rho=−0.47, P=0.001) (Fig 2A, Table 2). In multivariable analysis adjusting for age, sex, and fasting triglycerides, SM was no longer significant at P<0.05, while Cer(d18:1), Cer(d18:2), and to a lesser extent DHCer, remained statistically significant (Table 2). At the level of individual sphingolipids species, the relationships between SM species were generally attenuated compared to the Cer(d18:1) and Cer(d18:2) (Fig 2B). After adjustment, the strongest associations were among species containing between 20 and 24 carbons in Cer(d18:1) or Cer(d18:2) (Fig 2B). These data demonstrate a significant link between serum ceramides and gut microbiota richness, and reveal unexpected importance of long chain ceramides in this association.
Figure 2.
A: Gene count vs. lipid subclasses. Darkness of points reflects weight given to observation in robust regression (full weight = black, down-weighted = more white). Regression line is robust regression fit with R function ‘rlm.’ Statistics are provided in table 2. B: Manhattan-like plot of significance for robust regressions. Model 1 = unadjusted, Model 2 = age and sex adjusted, Model 3 = Model 2 + triglycerides and total cholesterol adjusted. DHCer are labeled by their chemical structure Cer(d18:0). C: Change in gene richness with dietary intervention in high (white) and low (black) baseline gene richness defined in Cotillard et al. Data are mean ± SE. D: Log2 fold-change of each subclass between baseline and week 6 (left) and week 12 (right), **P<0.01 versus week 0, ††P<0.01 versus week 0 and week 6. E: Standardized β-coefficients (and 95% confidence intervals) from regression of the lagged change in lipid subclasses onto lagged change in gene richness, stratified by high and low gene richness.
Table 2.
Associations between gene richness and sphingolipid subclasses.
| SM | DHCer | Cer(d18:1) | Cer(d18:2) | |||||
| Correlation | Rho | P | Rho | P | Rho | P | Rho | P |
| −0.36 | 0.017 | −0.25 | 0.102 | −0.45 | 0.002 | −0.47 | <0.001 | |
| Regression | Std-β | P | Std-β | P | Std-β | P | Std-β | P |
| - Model 1 | −0.52 | 0.018 | −0.29 | 0.018 | −0.46 | <0.001 | −0.53 | <0.001 |
| - Model 2 | −0.52 | 0.128 | −0.29 | 0.017 | −0.45 | <0.001 | −0.51 | <0.001 |
| - Model 3 | −0.53 | 0.054 | −0.23 | 0.033 | −0.34 | 0.003 | −0.37 | <0.001 |
Associations between gene count and lipid subclasses tested by Spearman correlation and robust regression. Model 1 = unadjusted, Model 2 = age and sex adjusted, Model 3 = Model 2 + triglycerides adjusted. Std-β = standardized coefficient.
Longitudinal changes in gene richness would bolster the cross-sectional findings. As reported previously (Cotillard et al. 2013), diet-induced weight loss significantly increased gene richness in subjects with low gene richness before the intervention (Fig 2C). We therefore examined if weight loss also corresponded to changes in circulating sphingolipids. DHCer, Cer(d18:2), Cer(d18:1), and SM were decreased by 47%, 39%, 35%, and 26%, respectively, after a 6-week period of diet restriction in the obese subjects (all P<0.01), and remained suppressed—with some rebound—during the 6 additional weeks of weight stabilization (Figure 2D). Over the weight loss period, few changes were seen in patients with high gene richness at baseline, whereas whose with initial low gene counts ameliorated their low microbiome diversity (Fig 2C). Regression of lagged variables confirmed a longitudinal association: an increase in gene richness from the previous time point was associated with a concomitant decrease in sphingolipid subclasses among participants with low baseline richness (Figure 2E). Overall, these data demonstrate association between blood ceramide levels and dynamic gut microbiota status driven by diet interventions in subjects with overweight/obesity.
3.3. Relationships between serum ceramides and specific metagenomic species (MGS)
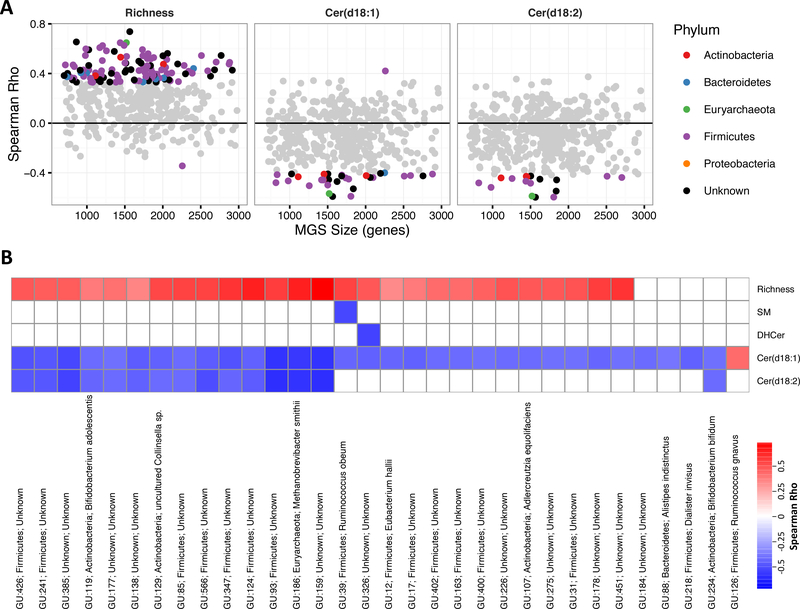
We next investigated if some specific bacterial species could be related to host ceramide levels. Lower gene richness was associated with a decrease in 97 of the 310 MGS (Padj<0.1; Fig 3A), consistent with the loss of bacterial richness. Firmicutes, the most abundant phylum in the human intestine, comprised the majority of associations with both gene richness and sphingolipids (Figure 3A). Decreased abundance of 30 MGS were associated with an increase serum concentrations of Cer(d18:1); approximately half of these bacteria (14 MGS) also showed significant association with Cer(d18:2) in the same direction (Figure 3B). A number of the sphingolipid-related MGS could be annotated to the species level. In particular, Methanobrevibacter smithii, Bifidobacterium adolescentis, Alderceutzia equolifaciens, Ruminococcus obeum, and Eubacteria hallii were associated with both low gene richness and high serum levels of Cer(d18:1). Alistipes indistinctus, Dialister invisus, and Bifidobacterium bifidum were also negatively associated with Cer(d18:1), but not with gene richness. Only one MGS was inversely associated with DHCer and one with SM. Ruminococcus gnavus was positively associated with Cer(d18:1). Thus from this analysis, it appears that the link between serum ceramide levels and gut microbe communities mainly relate to the loss of bacterial subgroups.
Figure 3.
A: Spearman correlations between lipid subclasses and each MGS plotted against the gene size of each MGS. Colored points are Padj<0.1 and reflect taxonomic annotation at the phylum level. B: Spearman correlations between each MGS and gene richness or lipid lipid subclass. Many more MGS were related to gene richness, therefore the heatmap is restricted to only MGS associated with Cer(d18:1) (Padj<0.1). Labels are “MGS id; phylum; species” (or genus if species undetermined).
3.4. Functional characterization of metagenomes associated with ceramide levels
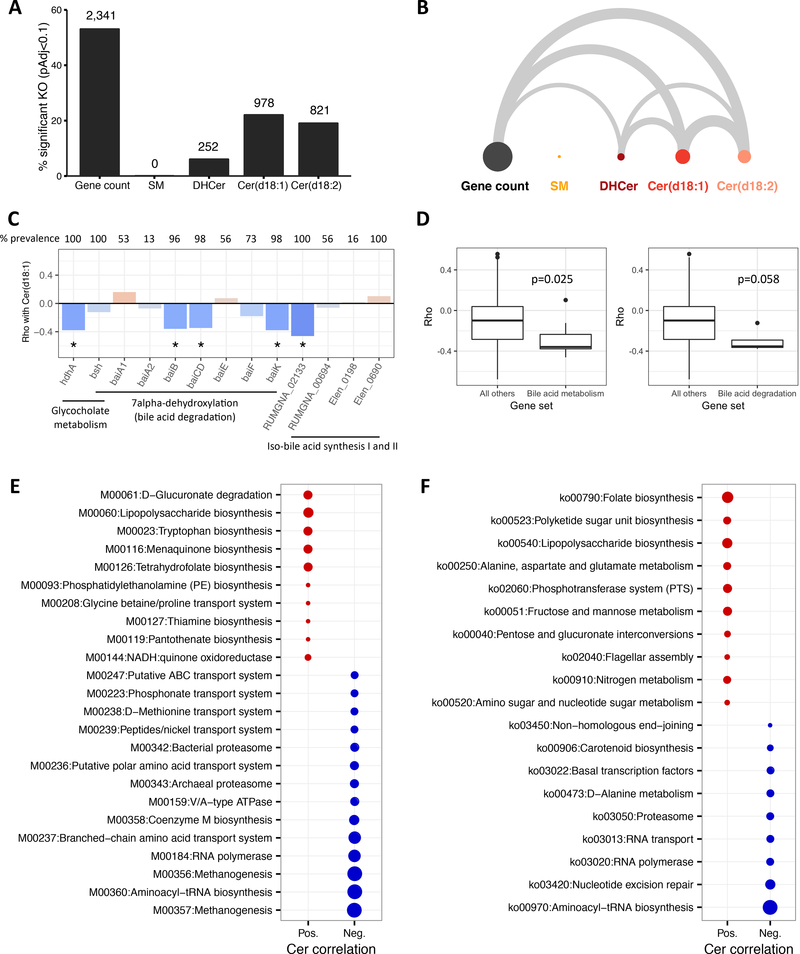
Quantitative metagenomics allows direct characterization of the functional potential of the microbiota. Consistent with the MGS data, gene richness was associated with changes in a large number of orthologue genes grouped within KO annotations (approximately half of total KOs detected). Interestingly, serum levels of Cer(d18:1) and Cer(d18:2) showed significant association with approximately 20% of annotated KO, and only a small percentage related to DHCer and no KOs significantly correlated to SM (Figure 4A). Ninety-one percent of Cer(d18:1) and 95% of Cer(d18:2) associated KOs were also associated with gene richness (Figure 4B).
Figure 4.
A: Percentage of total KOs significantly correlated (Padj<0.1) with gene count and lipid subclasses (number of significant KOs is above bars). B: Arc diagram where width of edge is proportional to the significant KOs shared between gene count and lipid subclasses. C. Spearman correlations between bile acid metabolism genes and Cer(d18:1), prevalence is percent of participants with abundance greater than 0, *Padj<0.1. D: Comparison of correlations between all bile acid metabolism genes (left) or bile acid degradation genes (right) and all KOs not in the respective gene sets, p-values are from the reporter features algorithm. E: Reporter modules positively (red) or negatively (blue) correlated to Cer(d18:1) (Padj<0.05). Circle size is proportional to statistical significance. F: Reporter pathways for Cer(d18:1) (Padj<0.05).
Because studies in mice have uncovered secondary bile acid metabolism by the gut microbiota as an important regulator of ceramides (Jiang et al. 2015), we examined the relationship between Cer(d18:1) and bacterial genes involved in bile acid transformation. Genes involved in glycoholate metabolism, bai genes coding the 7α-dehydroxylation pathway for secondary bile acid synthesis (bile acid degradation), and iso-bile acid synthesis genes were found with a decreased frequency with higher host serum ceramides levels (Padj<0.1), which suggest different potential for bile acid transformation in subjects with elevated ceramides. Analysis at the module level confirmed the inverse association with bile acid metabolism genes (p=0.025), and bile acid degradation (P=0.058) more specifically (Figure 4D). Longitudinal data confirmed these findings since patients with low richness at baseline exhibited an increase in abundance of several bile acid genes, including bsh, during the dietary intervention concomitant with the increase in richness and decrease in ceramides (Online Resource 4). A hypothesis-free analysis of all available KEGG modules and pathways was also performed at baseline. At the module level (Figure 4E), the strongest inverse association with serum Cer(d18:1) were related to methanogenesis and cellular processes comprised of archaea-specific enzymes (RNA polymerase and archaeal proteasome). Lower capacity for nutrient transport including branched-chain amino acids and polar amino acids were a notable signature bacterial communities with elevated host Cer(d18:1). At the pathway level (Figure 4F), lower abundance of general cellular processes, metabolism of alanine, and carotenoid biosynthesis were detected. The majority of KEGG modules enriched with higher Cer(d18:1) were related to biosynthesis. Lipopolysaccharide biosynthesis was highly enriched at the module level, as was the biosynthesis of amino acids (tryptophan) and B-vitamins (thiamine, pantothenate). Higher capacity for LPS biosynthesis and flagellar assembly were positively related to Cer(d18:1) at the pathway level. Thus, differences in overall metagenomic richness, bacterial abundance, and functional capacity are related to host ceramides accumulation.
4. Discussion
We herein establish an association between the human gut microbiota and ceramides—lipid mediators implicated in the development of type 2 diabetes. After confirming the links between serum ceramides and diabetes risk, we found that low metagenomic richness was associated with significantly higher levels of serum ceramides. Decreased abundance of Methanobreviibacter smithii and anti-inflammatory bifidobacteria species were related to higher ceramides levels. Importantly, the ceramide-associated microbiome demonstrated an increase in a number of biosynthetic pathways, notably increased LPS synthesis and flagellar assembly. Thus, low metagenomic richness may contribute to increased diabetes risk in humans by permitting an inflammatory dysbiosis that induces ceramide accumulation.
Studies in mice have implicated the gut microbiota in the regulation of host metabolism (Turnbaugh et al. 2006). Gut microbiota composition is also altered in patients with type 2 diabetes from both China and Europe (Qin et al. 2012; Karlsson et al. 2013). In the Chinese population, the individuals with type 2 diabetes had an increase in a number of pathogenic bacteria and a decrease in anti-inflammatory bacteria (Qin et al. 2012). In Swedish individuals, type 2 diabetes and elevated triglycerides were primarily associated with decreased abundance of bacterial species, and their metagenomes were enriched in fructose and mannose metabolism and phosphotransferase systems, similar to our findings in subjects without type 2 diabetes. More recently, it was shown that metformin treatment at least partially confounds the relationship with type 2 diabetes (Forslund et al. 2015), therefore a major strength of the current study is the persistence of these findings in untreated participants. Decreased gut microbial richness is observed in overweight and obese patients, and is associated with markers of insulin resistance, systemic inflammation (e.g. CRP), and hypertriglyceridemia (Le Chatelier et al. 2013), and these factors are improved with diet-induced weight loss (Cotillard et al. 2013). Low gene richness has also been associated with more severe conditions such as ulcerative colitis (Lepage et al. 2011). Thus, microbiota richness is lower with both metainflammation and chronic inflammatory diseases, which may stimulate ceramide production.
In addition to lipid loading, toll-like receptor activation leads to the accumulation of ceramides (Chaurasia & Summers 2015). Lipopolysaccharide, the major TLR-4 ligand, induces both de novo ceramide synthesis (Memon et al. 1998) and conversion of SM into ceramides via sphingomyelinases (Sakata et al. 2007). Endotoxin-induced cytokines also lead to ceramide accumulation (Memon et al. 1998). LPS can induce ceramide accumulation in muscle and liver (Holland et al. 2011), as well as lead to increased secretion of ceramides into lipoprotein particles and serum (Memon et al. 1998). Chronic endotoxemia and cytokine signaling could therefore induce ceramide accumulation in obesity. Indeed, high fat feeding in mice increases circulating LPS two- to three-fold and increases the proportion of LPS-containing bacteria in the gut (Cani et al. 2007a). Conversely, prebiotic treatment that restores bifidobacterium species in the intestine of rodents are correlated with reduced endotoxemia and improved glucose homeostasis (Cani et al. 2007b). We observed a decrease in Bifidobacterium bifidum and Bifidobacerium adolescentis with higher ceramide levels, consistent with these observations in mice. Notably, Ruminococcus gnavus was positively associated with ceramides, and this species is increased in inflammatory bowel disease and appears to regulate the composition of the mucosa-associated bacterial network in these diseases (Png et al. 2010). The positive associations between increased LPS biosynthesis and flagellar assembly, another pathogen-associated molecular pattern (Hayashi et al. 2001), corroborates the link between ceramides and gut-derived inflammation. Together, albeit only observational, our results support the hypothesis that the loss of overall diversity permits a shift towards increased endotoxin burden in the intestine, activating inflammatory pathways in the host, i.e. ceramide accumulation.
Microbial bile acid metabolism may also regulate host ceramides synthesis. Jiang et al. showed that the gut microbiota regulates serum ceramide levels through intestinal farsenoid-X-receptor (FXR) signaling (Jiang et al. 2015). Specifically, they found that reduced hydrolysis of tauro-beta-muricholic acid (by bsh), an antagonist of intestinal FXR signaling, reduces intestinal FXR activation, mitochondrial dysfunction, and circulating ceramides. In contrast, we did not detect a relationship between bsh gene abundance, and even found that reduced capacity for overall secondary bile acid metabolism was associated with higher circulating ceramides. The design of our study prohibits conclusive statements regarding the role of secondary bile acid metabolism and ceramides, but future studies incorporating careful analysis of fecal and serum bile acid levels, as well as bsh activity in stool, could help clarify this potentially important mechanism.
Regarding different ceramide species, we found that longer chain Cer(d18:1), namely C20, C22, and C24 species, were the most strongly associated with both metabolic parameters and gene richness. This acyl distribution, regulated by the CERS2 gene, suggests that the gut microbiota may be particularly related to hepatic ceramide synthesis (Levy & Futerman 2010). These results are contrary to recent genetic studies in mice that show that Cer(d18:1/16:0) is the primary driver of metabolic dysfunction (Turpin et al. 2014), and other reports of no effect (Raichur et al. 2014) or even a beneficial effect of very-long chain ceramides (Montgomery et al. 2016). Similar conclusions were also drawn from an epidemiological study of coronary artery disease (Laaksonen et al. 2016). On the other hand, nearly all circulating ceramides, regardless of fatty acids, have approximately the same degree of association with type 2 diabetes and insulin resistance (Haus et al. 2009), or, as with our study, show equivalent or greater elevations in C18 to C24 ceramides compared to Cer(d18:1/16:0) (Meikle et al. 2013; Shui et al. 2013). Consistent with the relationships to β-cell function in our study, circulating very-long chain DHCer predict incident type 2 diabetes (Wigger et al. 2017). These discrepant findings may be reconciled by a specific action of circulating ceramides. LDL-enriched in Cer(d18:1/16:0) or Cer(d18:1/24:0), the latter being far more abundant in serum, induce insulin resistance equivalently in vitro and in vivo (Boon et al. 2013). Further, extracellular ceramides, derived from exogenous delivery, can enhance hepatic gluconeogenesis, consistent with the positive association with fasting glucose in our study (Xie et al. 2017). Further work is therefore needed to determine the causal role, if any, of circulating ceramides, and their acyl-chain length, in the pathogenesis of insulin resistance in humans.
Several limitations of our study should be noted. Most of the evidence for ceramide-induced insulin resistance comes from cellular accumulation within metabolic tissues. While we have discussed the evidence for a potential role for circulating ceramides, future studies that include both metagenomic analysis and tissue lipidomics would be an important next step. Our study is observational: while we have adjusted for a number of potentially important confounders, larger human studies and gut microbiota transfer studies would both aid in confirming the importance of the gut microbiota on ceramide metabolism.
In conclusion, we confirmed that serum ceramides are positively correlated with markers of both insulin resistance and impaired β-cell function. Inflammation is an important driver of ceramide synthesis, and our data suggest that gut dysbiosis, characterized by decreased metagenomic richness, lower abundance of anti-inflammatory bifidobacteria species, and a greater metagenomic capacity for toll-like receptor ligand synthesis, may contribute to this pathway in human obesity. This study paves the way for future experiments aimed at elucidating the role of the gut microbiota in host lipid metabolism and the pathogenesis of type 2 diabetes.
Supplementary Material
Online resource 4. Fold change (on log base 2 scale) of each bile acid gene between baseline and weeks 6 and 12, stratified by baseline gene richness. Prevalence is number of metagenome samples (subject 1 at baseline is a different sample from subject 1 at week 6) with abundance greater than 0. Statistical significance was determined by paired Wilcoxon tests with *P<0.05 and **P<0.01. Fold-change of means was calculated because too many zeros were present in some strata to calculate fold-change (resulted in infinity or undefined ratios).
Online resource 3. Spearman correlations between sphingolipids and clinical variables. *P<0.05, **P<0.01, ***P<0.001. % BF = Percent body fat, AdipoDia = adipocyte diameter, TG = fasting triglycerides, NEFA = non-esterified free fatty acids, sCD14 = soluble CD14, OGTT-60 and OGTT-120 = plasma glucose during OGTT at 60 and 120 minutes, respectively, ISSI-2 = insulin-secretion-sensitivity-index-2. DHCer are labeled by their chemical structure Cer(d18:0).
Acknowledgments
Compliance with ethical standards
The study was authorized by the Ethical Committee (CPP N° 1 Hôtel Dieu Hospital) and all procedures were performed in accordance with ethical and institutional standards. The study is registered on clinicaltrials.gov: NCT01314690.
Acknowledgements
Thank you to Sophie Gougis, Soraya Fellahi (Dept. of Biochemistry and Hormonology, Tenon hospital), Dominique Bonnefont-Rousselot and Randa Bittar (Department of Metabolic Biochemistry, Pitié-Salpêtrière hospital) for their contributions to data collection and analysis. We would also like to thank the nurses and technicians, and of course, the patients themselves for their invaluable contribution.
Funding
This work was supported by Agence Nationale de la Recherche (ANR MICRO-Obes), KOT-Ceprodi and the association Foundation Coeur et Arteres (clinical investigation) as well as European Union’s Seventh Framework Program Metacardis under grant agreement HEALTH-F4–2012-305312 and Horizon 2020 Framework Program (EPoS, grant #634413). The European Foundation for the Study of Diabetes Albert Renold travel fellowship have contributed to the work of BDK and Danone Research to the work of MCD and KC.
Footnotes
Informed consent
Informed consent was obtained from all individual participants included in the study.
Self-Archiving
This is a post-peer-review, pre-copyedit version of an article published in Metabolomics. The final authenticated version is available online at: http://dx.doi.org/10.1007/s11306-019-1596-0.
MICRO-Obes consortium
Aurélie Cotillard; Sean P Kennedy; Nicolas Pons; Emmanuelle Le Chatelier; Mathieu Almeida; Benoit Quinquis; Nathalie Galleron; Jean-Michel Batto; Pierre Renault; Stanislav Dusko Ehrlich; Hervé Blottière; Marion Leclerc; Tomas de Wouters; Patricia Lepage, Joel Doré
Conflict of interest
All authors declare that they have no conflicts of interest.
Data availability state
SOLiD sequencing data have been deposited in the European Bioinformatics Institute (EBI) European Nucleotide Archive (ENA) under accession number ERP003699.
References
- Antuna-Puente B, Disse E, Faraj M, Lavoie ME, Laville M, Rabasa-Lhoret R et al. (2009) Evaluation of insulin sensitivity with a new lipid-based index in non-diabetic postmenopausal overweight and obese women before and after a weight loss intervention. Eur J Endocrinol 161: 51–56. [DOI] [PubMed] [Google Scholar]
- Aron-Wisnewsky J, Tordjman J, Poitou C, Darakhshan F, Hugol D, Basdevant A et al. (2009) Human adipose tissue macrophages: m1 and m2 cell surface markers in subcutaneous and omental depots and after weight loss. J Clin Endocrinol Metab 94: 4619–4623. [DOI] [PubMed] [Google Scholar]
- Boon J, Hoy AJ, Stark R, Brown RD, Meex RC, Henstridge DC et al. (2013) Ceramides contained in LDL are elevated in type 2 diabetes and promote inflammation and skeletal muscle insulin resistance. Diabetes 62: 401–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camont L, Lhomme M, Rached F, Le Goff W, Nègre-Salvayre A, Salvayre R et al. (2013) Small, Dense High-Density Lipoprotein-3 Particles Are Enriched in Negatively Charged Phospholipids Relevance to Cellular Cholesterol Efflux, Antioxidative, Antithrombotic, Anti-Inflammatory, and Antiapoptotic Functionalities. Arteriosclerosis, thrombosis, and vascular biology 33: 2715–2723. [DOI] [PubMed] [Google Scholar]
- Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D et al. (2007a) Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 56: 1761–1772. [DOI] [PubMed] [Google Scholar]
- Cani PD, Neyrinck AM, Fava F, Knauf C, Burcelin RG, Tuohy KM et al. (2007b) Selective increases of bifidobacteria in gut microflora improve high-fat-diet-induced diabetes in mice through a mechanism associated with endotoxaemia. Diabetologia 50: 2374–2383. [DOI] [PubMed] [Google Scholar]
- Chaurasia B & Summers SA (2015) Ceramides - Lipotoxic Inducers of Metabolic Disorders. Trends Endocrinol Metab 26: 538–550. [DOI] [PubMed] [Google Scholar]
- Clemente JC, Pehrsson EC, Blaser MJ, Sandhu K, Gao Z, Wang B et al. (2015) The microbiome of uncontacted Amerindians. Science Advances 1: e1500183–e1500183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotillard A, Kennedy SP, Kong LC, Prifti E, Pons N, Le Chatelier E et al. (2013) Dietary intervention impact on gut microbial gene richness. Nature 500: 585–588. [DOI] [PubMed] [Google Scholar]
- Forslund K, Hildebrand F, Nielsen T, Falony G, Le Chatelier E, Sunagawa S et al. (2015) Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 528: 262–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass CK & Olefsky JM (2012) Inflammation and lipid signaling in the etiology of insulin resistance. Cell Metab 15: 635–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haus JM, Kashyap SR, Kasumov T, Zhang R, Kelly KR, DeFronzo RA et al. (2009) Plasma ceramides are elevated in obese subjects with type 2 diabetes and correlate with the severity of insulin resistance. Diabetes 58: 337–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi F, Smith KD, Ozinsky A, Hawn TR & Eugene… C.Y. (2001) The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature [DOI] [PubMed] [Google Scholar]
- Holland WL, Bikman BT, Wang LP, Yuguang G, Sargent KM, Bulchand S et al. (2011) Lipid-induced insulin resistance mediated by the proinflammatory receptor TLR4 requires saturated fatty acid-induced ceramide biosynthesis in mice. J Clin Invest 121: 1858–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Xie C, Li F, Zhang L, Nichols RG, Krausz KW et al. (2015) Intestinal farnesoid X receptor signaling promotes nonalcoholic fatty liver disease. J Clin Invest 125: 386–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson FH, Tremaroli V, Nookaew I, Bergstrom G, Behre CJ, Fagerberg B et al. (2013) Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 498: 99–103. [DOI] [PubMed] [Google Scholar]
- Katz A, Nambi SS, Mather K & Baron AD (2000) Quantitative insulin sensitivity check index: a simple, accurate method for assessing insulin sensitivity in humans. The Journal of … [DOI] [PubMed] [Google Scholar]
- Kong LC, Holmes BA, Cotillard A, Habi-Rachedi F, Brazeilles R, Gougis S et al. (2014) Dietary patterns differently associate with inflammation and gut microbiota in overweight and obese subjects. PLoS One 9: e109434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laaksonen R, Ekroos K, Sysi-Aho M, Hilvo M, Vihervaara T, Kauhanen D et al. (2016) Plasma ceramides predict cardiovascular death in patients with stable coronary artery disease and acute coronary syndromes beyond LDL-cholesterol. Eur Heart J 37: 1967–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G et al. (2013) Richness of human gut microbiome correlates with metabolic markers. Nature 500: 541–546. [DOI] [PubMed] [Google Scholar]
- Lepage P, Häsler R, Spehlmann ME, Rehman A, Zvirbliene A, Begun A et al. (2011) Twin study indicates loss of interaction between microbiota and mucosa of patients with ulcerative colitis. Gastroenterology 141: 227–236. [DOI] [PubMed] [Google Scholar]
- Levy M & Futerman AH (2010) Mammalian ceramide synthases. IUBMB Life NA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupi R, Dotta F, Marselli L, Del Guerra S, Masini M, Santangelo C et al. (2002) Prolonged exposure to free fatty acids has cytostatic and pro-apoptotic effects on human pancreatic islets: evidence that beta-cell death is caspase mediated, partially dependent on ceramide pathway, and Bcl-2 regulated. Diabetes 51: 1437–1442. [DOI] [PubMed] [Google Scholar]
- Matsuda M & DeFronzo RA (1999) Insulin sensitivity indices obtained from oral glucose tolerance testing: comparison with the euglycemic insulin clamp. Diabetes care 22: 1462–1470. [DOI] [PubMed] [Google Scholar]
- Meikle PJ, Wong G, Barlow CK, Weir JM, Greeve MA, MacIntosh GL et al. (2013) Plasma lipid profiling shows similar associations with prediabetes and type 2 diabetes. PLoS One 8: e74341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memon RA, Holleran WM, Moser AH, Seki T, Uchida Y, Fuller J et al. (1998) Endotoxin and Cytokines Increase Hepatic Sphingolipid Biosynthesis and Produce Lipoproteins Enriched in Ceramides and Sphingomyelin. Arteriosclerosis, Thrombosis, and Vascular Biology 18: 1257–1265. [DOI] [PubMed] [Google Scholar]
- Montgomery MK, Brown SH, Lim XY, Fiveash CE, Osborne B, Bentley NL et al. (2016) Regulation of glucose homeostasis and insulin action by ceramide acyl-chain length: A beneficial role for very long-chain sphingolipid species. Biochim Biophys Acta 1861: 1828–1839. [DOI] [PubMed] [Google Scholar]
- Nielsen HB, Almeida M, Juncker AS, Rasmussen S, Li J, Sunagawa S et al. (2014) Identification and assembly of genomes and genetic elements in complex metagenomic samples without using reference genomes. Nat Biotechnol 32: 822–828. [DOI] [PubMed] [Google Scholar]
- Png CW, Lindén SK, Gilshenan KS, Zoetendal EG, McSweeney CS, Sly LI et al. (2010) Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol 105: 2420–2428. [DOI] [PubMed] [Google Scholar]
- Prifti E, Chatelier EL, Pons N, Almeida M, Leonard P, Batto JM et al. (2013) MetaOMineR: A fine-tuned pipeline for whole metagenomic data analyses. International Human Microbiome Congress (4th) [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C et al. (2010) A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464: 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F et al. (2012) A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490: 55–60. [DOI] [PubMed] [Google Scholar]
- Qin N, Yang F, Li A, Prifti E, Chen Y, Shao L et al. (2014) Alterations of the human gut microbiome in liver cirrhosis. Nature 513: 59–64. [DOI] [PubMed] [Google Scholar]
- Raichur S, Wang ST, Chan PW, Li Y, Ching J, Chaurasia B et al. (2014) CerS2 haploinsufficiency inhibits beta-oxidation and confers susceptibility to diet-induced steatohepatitis and insulin resistance. Cell Metab 20: 687–695. [DOI] [PubMed] [Google Scholar]
- Retnakaran R, Qi Y, Goran MI & Hamilton JK (2009) Evaluation of proposed oral disposition index measures in relation to the actual disposition index. Diabet Med 26: 1198–1203. [DOI] [PubMed] [Google Scholar]
- Retnakaran R, Shen S, Hanley AJ, Vuksan V, Hamilton JK & Zinman B (2008) Hyperbolic relationship between insulin secretion and sensitivity on oral glucose tolerance test. Obesity (Silver Spring) 16: 1901–1907. [DOI] [PubMed] [Google Scholar]
- Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL et al. (2013) Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 341: 1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizkalla SW, Prifti E, Cotillard A, Pelloux V, Rouault C, Allouche R et al. (2012) Differential effects of macronutrient content in 2 energy-restricted diets on cardiovascular risk factors and adipose tissue cell size in moderately obese individuals: a randomized controlled trial. Am J Clin Nutr 95: 49–63. [DOI] [PubMed] [Google Scholar]
- Sakata A, Ochiai T, Shimeno H, Hikishima S, Yokomatsu T, Shibuya S et al. (2007) Acid sphingomyelinase inhibition suppresses lipopolysaccharide-mediated release of inflammatory cytokines from macrophages and protects against disease pathology in dextran sulphate sodium-induced colitis in mice. Immunology 122: 54–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shui G, Lam SM, Stebbins J, Kusunoki J, Duan X, Li B et al. (2013) Polar lipid derangements in type 2 diabetes mellitus: potential pathological relevance of fatty acyl heterogeneity in sphingolipids. Metabolomics 9: 786–799. [Google Scholar]
- Stratford S, Hoehn KL, Liu F & Summers SA (2004) Regulation of insulin action by ceramide: dual mechanisms linking ceramide accumulation to the inhibition of Akt/protein kinase B. J Biol Chem 279: 36608–36615. [DOI] [PubMed] [Google Scholar]
- Summers SA (2018) Could Ceramides Become the New Cholesterol. Cell Metab 27: 276–280. [DOI] [PubMed] [Google Scholar]
- Tremaroli V, Karlsson F, Werling M, Ståhlman M, Kovatcheva-Datchary P, Olbers T et al. (2015) Roux-en-Y Gastric Bypass and Vertical Banded Gastroplasty Induce Long-Term Changes on the Human Gut Microbiome Contributing to Fat Mass Regulation. Cell Metab 22: 228–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER & Gordon JI (2006) An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444: 1027–1031. [DOI] [PubMed] [Google Scholar]
- Turpin SM, Nicholls HT, Willmes DM, Mourier A, Brodesser S, Wunderlich CM et al. (2014) Obesity-induced CerS6-dependent C16:0 ceramide production promotes weight gain and glucose intolerance. Cell Metab 20: 678–686. [DOI] [PubMed] [Google Scholar]
- Wallace TM, Levy JC & Matthews DR (2004) Use and abuse of HOMA modeling. Diabetes Care 27: 1487–1495. [DOI] [PubMed] [Google Scholar]
- Wigger L, Cruciani-Guglielmacci C, Nicolas A, Denom J, Fernandez N, Fumeron F et al. (2017) Plasma Dihydroceramides Are Diabetes Susceptibility Biomarker Candidates in Mice and Humans. Cell Rep 18: 2269–2279. [DOI] [PubMed] [Google Scholar]
- Xie C, Jiang C, Shi J, Gao X, Sun D, Sun L et al. (2017) An Intestinal Farnesoid X Receptor-Ceramide Signaling Axis Modulates Hepatic Gluconeogenesis in Mice. Diabetes 66: 613–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yohai VJ (1987) High breakdown-point and high efficiency robust estimates for regression. The Annals of Statistics [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Online resource 4. Fold change (on log base 2 scale) of each bile acid gene between baseline and weeks 6 and 12, stratified by baseline gene richness. Prevalence is number of metagenome samples (subject 1 at baseline is a different sample from subject 1 at week 6) with abundance greater than 0. Statistical significance was determined by paired Wilcoxon tests with *P<0.05 and **P<0.01. Fold-change of means was calculated because too many zeros were present in some strata to calculate fold-change (resulted in infinity or undefined ratios).
Online resource 3. Spearman correlations between sphingolipids and clinical variables. *P<0.05, **P<0.01, ***P<0.001. % BF = Percent body fat, AdipoDia = adipocyte diameter, TG = fasting triglycerides, NEFA = non-esterified free fatty acids, sCD14 = soluble CD14, OGTT-60 and OGTT-120 = plasma glucose during OGTT at 60 and 120 minutes, respectively, ISSI-2 = insulin-secretion-sensitivity-index-2. DHCer are labeled by their chemical structure Cer(d18:0).