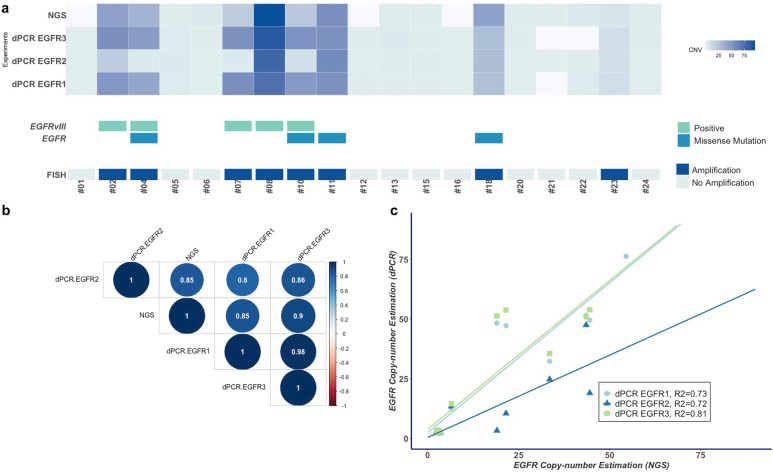
Fig. 2.
Concordance between the results of the dPCR assay, FISH, next-generation sequencing (NGS) and LD-RT-PCR for the detection of the EGFR amplification and EGFRvIII variant in the exploratory cohort (n = 19). a Heatmap of EGFR copy number estimated by NGS and the three dPCR assays. Each column represents a tumor sample (n = 19). The blue gradient represents the estimated value of the EGFR copy number. There is a strong agreement between the EGFR1 and EGFR2 dPCR assays and NGS. The absence of results using the NGS experiment is indicated by the light gray color. Below the heatmap, the presence of EGFR mutations and EGFRvIII variant as well as the results of FISH are presented. The presence of somatic mutations was detected by the EGFR-targeted NGS panel, and the presence of the EGFRvIII variant was detected by LD-RT-PCR. Patient #08 harbors both the EGFR amplification and EGFRvIII variant with tumor heterogeneity regarding the copy number estimation by dPCR (EGFR1 63, EGFR2 70 and EGFR3 91). b Correlation matrix plot of EGFR copy number estimation using three dPCR assays (EGFR1, EGFR2 and EGFR3) and NGS (n = 16). The dPCR EGFR3 assay results have the highest correlation with the NGS results. On the other hand, the dPCR EGFR2 assay results have the poorest correlation, mainly due to its underestimation of the EGFR CNV in the case of EGFRvIII-positive glioblastoma. c Linear regression curves representing EGFR copy number values estimated by NGS (x-axis) and the copy number estimated by the three dPCR assays (y-axis). As expected with the results of the matrix correlation plot, the estimation using the dPCR EGFR3 assay was confirmed to have the best correlation to the NGS estimation