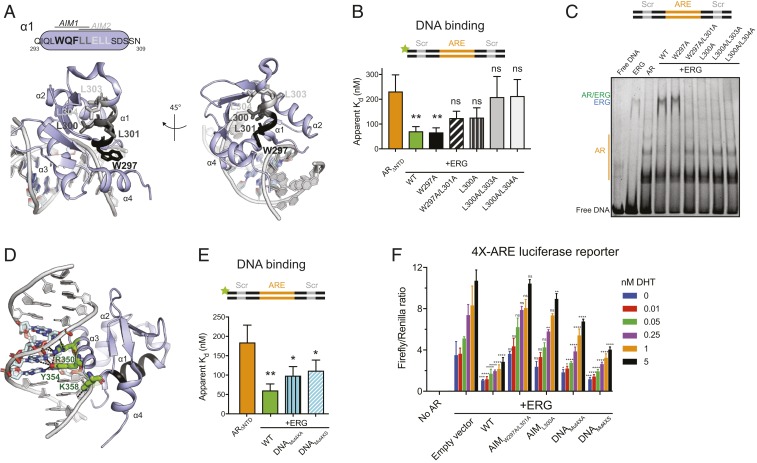
Fig. 3.
Identification of residues important for ERG-mediated stimulation of AR activity. (A) Mapping of AIMs onto helix 1 in the crystal structure of ERG bound to ETS dsDNA (Protein Data Bank [PDB]: 4IRI). Candidate AR-interacting residues are shown as sticks. Black residues correspond to putative AIM1, and light gray AIM2; dark gray is common to both motifs. This and other structure figures were generated with PyMol. (B) AR binding to fluorescein-labeled ARE/Scr DNA in the presence of WT and AIM mutant ERG. (C) EMSA of ARE/Scr dsDNA in the presence of ARΔNTD and WT and ERG variants. Gel (4 to 20% TBE PAGE) is stained with Sybr Gold. (D) ERG DNA binding residues targeted for mutagenesis (green sticks) mapped onto the structure in A. (E) AR binding to fluorescein-labeled ARE/Scr DNA in the presence of WT and DNA binding mutant ERG variants. Data in B and E were acquired by fluorescence polarization and are shown as mean ± SD (n = 3 technical replicates; one-way ANOVA). (F) Activation of a 4×-ARE firefly luciferase reporter by AR-VP16 and regulation of AR by WT and ERG mutants in HEK293 cells. Data were acquired 24 h after transfection and normalized to a control Renilla luciferase reporter. Data are shown as mean ± SD (n = 3 biological replicates; two-way ANOVA). ****P ≤ 0.0001; ***P ≤ 0.001; **P ≤ 0.01; *P ≤ 0.05; ns, not significant, P > 0.05.