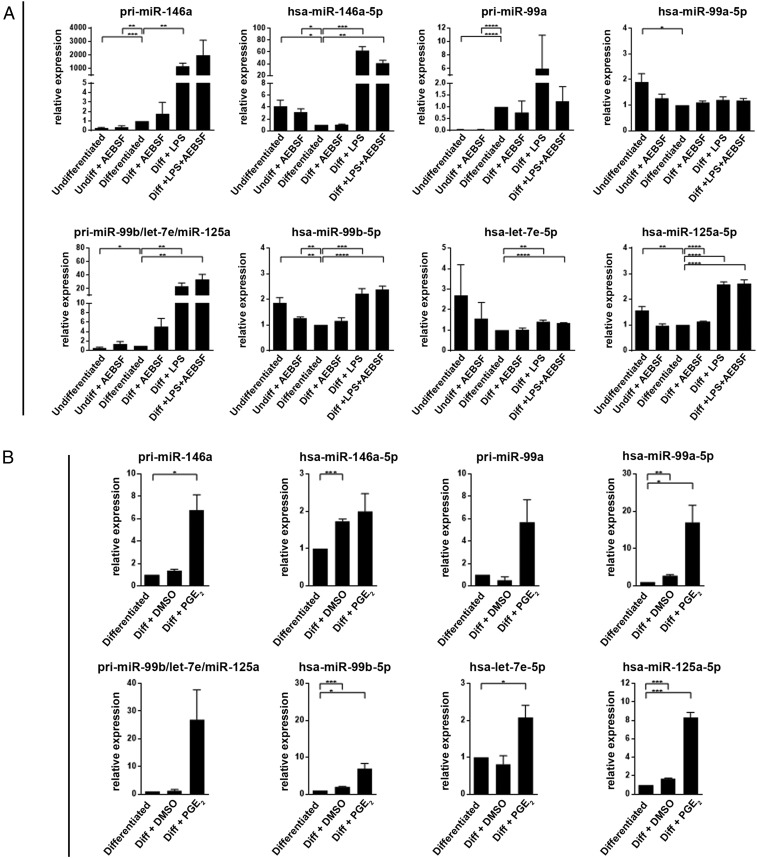
Fig. 6.
qPCR analysis of miRNAs in MM6 cells. (A) Effects of LPS and AEBSF. MM6 cells were differentiated (Diff) with TGF-β + 1,25diOHvitD3 for 96 h. LPS (1 µg/mL) was added as indicated at time 72 h (present final 24 h). AEBSF (150 to 250 µM) was added at time 80 h (present final 16 h). Performed with the Qiagen miScript Primer Assay (Methods). The data are presented relative to cells differentiated with only TGF-β+ 1,25diOHvitD3, set as 1. For the pri-miRNA, GAPDH was reference gene. For the mature miRNA, U6 was reference gene. Mean ± SEM, n = 3 to 4, two-tailed unpaired t test, ****P < 0.0001; ***P < 0.001; **P < 0.01; *P < 0.05. (B) Effect of PGE2. MM6 cells were differentiated with TGF-β + 1,25diOHvitD3 in presence of PGE2 (5 µM) or vehicle (DMSO) added at time 0 of the 96-h differentiation period. The data are presented relative to cells differentiated with only TGF-β + 1,25diOH, set as 1. For the pri-miRNA, GAPDH was reference gene. For the mature miRNA, U6 was reference gene. Mean ± SEM, n = 3, two-tailed unpaired t test, ***P < 0.001; **P < 0.01; *P < 0.05.