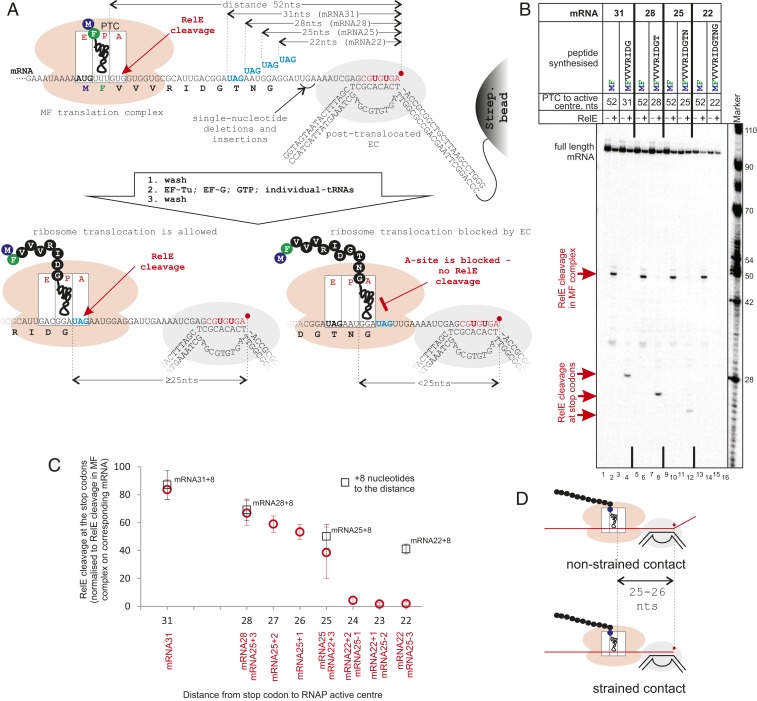
Fig. 2.
The distance between the coupled ribosome and RNAP interacting on mRNA. (A and B) Scheme and results of the experiment. MF complexes are coupled to ECs as in Fig. 1A, except here the EC is walked to the abasic position, where it is stabilized in the posttranslocated state (SI Appendix, Fig. S3A) and cannot extend mRNA further. mRNAs contain UAG stop codons (to standardize efficiency by RelE cleavage among mRNAs) at different distances from the RNAP active center. Ribosomes translate 9-, 10-, 11-, or 12-aa peptides toward the EC before they reach UAG stop codons. The availability of UAG in the A-site, as a measure of successful translocation of the ribosome, is tested by RelE cleavage (lanes 4, 8, 12, and 16). Ribosomes that came in contact with RNAP and cannot translocate have peptidyl-tRNA in the A-site, making them inaccessible for RelE (lane 16). (C) Efficiencies of RelE cleavage at UAGs located at various distances from the RNAP active center. The positions of one, two, and three nucleotide deletions and insertions in mRNAs are shown in A and in SI Appendix, Fig. S1A. Data points are means and error bars are SDs from 3 to 13 experiments. Black squares are the RelE cleavage efficiencies on the corresponding mRNAs when the EC was walked 8 bp (on a template without abasic site) further away from stop codons. (D) The distances on mRNA between the RNAP active center and the ribosome’s PTC in both the “strained” contact, when the ribosome is forced into a stalled EC (Bottom), and the “nonstrained” contact, when ribosome translocation pushes the backtracked RNAP (Top) are the same, 25 to 26 nucleotides.