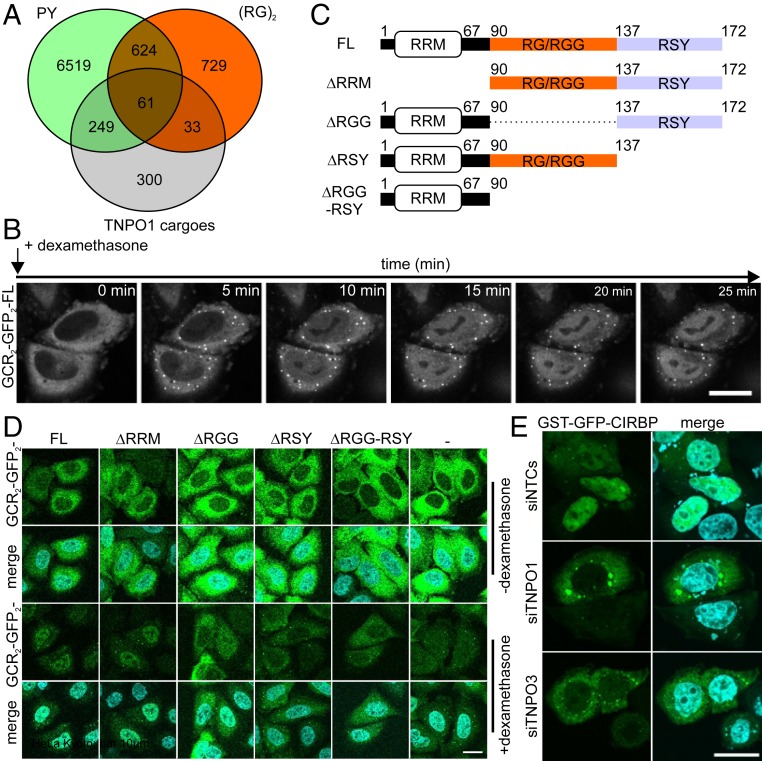
Fig. 1.
The RG/RGG- and RSY-rich regions in CIRBP mediate TNPO1- and 3-dependent nuclear import in HeLa cells. (A) Venn diagram corresponding to the PROSITE analysis (https://prosite.expasy.org/scanprosite/) of two motifs against the nonredundant protein sequences database filtered for human proteins (taxid:9606) harboring either 1) a di-RG motif (orange) each spaced by zero to five amino acids [R-G-x(0-5)-R-G] or 2) a PY motif (green) preceded by a basic amino acid spaced by zero to nine residues [[RKH]-x(0,9)-P-Y]. These two groups of proteins were compared with the published list of TNPO1 cargoes identified by Kimura et al. (16) and Mackmull et al. (17). (B) Time-dependent nuclear import of GCR2-GFP2-CIRBP full length in HeLa Kyoto cells on addition of dexamethasone visualized by live cell imaging. (Scale bar: 20 µm.) (C) Scheme of CIRBP illustrating its domain organization and deletion of individual domains for analysis of their involvement in nuclear import of CIRBP. (D) HeLa Kyoto cells were transiently transfected with constructs coding for GCR2-GFP2-CIRBP full length (FL) or CIRBP mutants lacking either the RRM (DRRM), the RG/RGG (DRGG), the RSY (DRSY), or both RG/RGG and RSY regions (DRGG-RSY). A construct coding for only GCR2-GFP2 (−) was used as control for diffusion. Cells were either fixed before (−dexamethasone) or after (+dexamethasone) the addition of 5 µM dexamethasone for 15 min at 37 °C. Nuclei were stained with DAPI, and cells were analyzed by fluorescence microscopy. (Scale bar: 10 µm.) (E) HeLa Kyoto cells transfected with either control small inhibitory RNA (siRNA) or siRNA against TNPO1 or TNPO3, respectively, were transfected with a construct coding for GST-GFP-CIRBP. Cells were fixed, nuclei were counterstained with DAPI, and cells were analyzed by fluorescence microscopy. (Scale bar: 20 µm.)