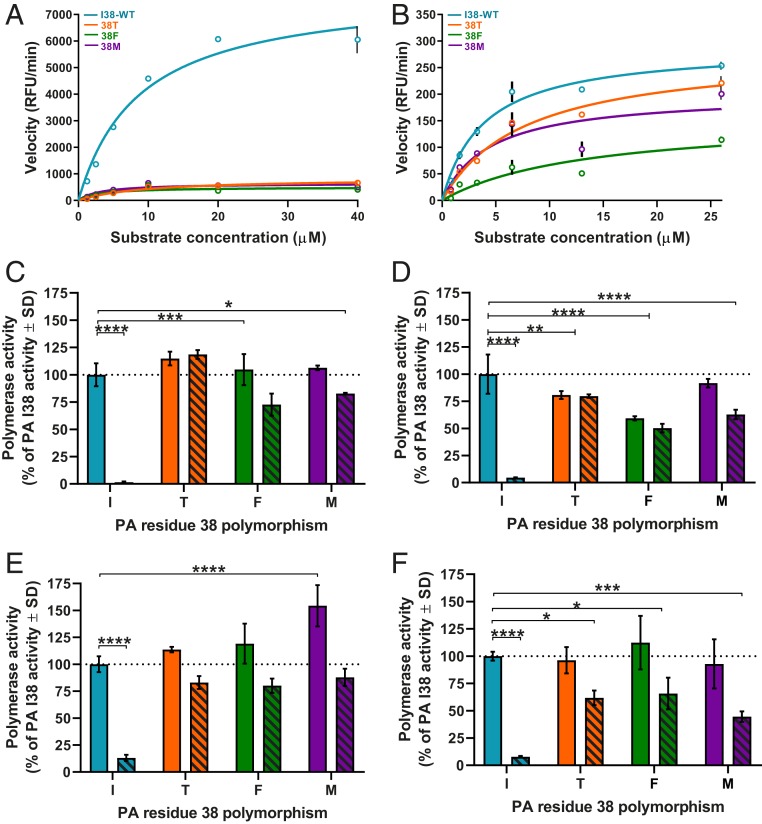
Fig. 1.
Endonuclease and minireplicon polymerase complex activity of I38-WT and 38T/F/M. The rPAN (5 µg) from (A) A/CA/04 (H1N1)pdm09 or (B) B/BR/60 (Victoria lineage) with a single amino acid substitution at residue 38 of the PA protein (I38-WT and 38T/F/M) was incubated with fluorescently labeled ssDNA substrate. PAN enzymatic activity and reaction rates were determined from measured release of fluorescence upon substrate cleavage over time (RFU/min). Polymerase activity of (C) A/CA/04 (H1N1)pdm09, (D) A/TX/71 (H3N2), (E) B/BR/60 (Victoria lineage), or (F) B/PH/3073 (Yamagata lineage) was determined by minireplicon assay in HEK293T cells transfected with plasmids expressing virus proteins NP, PB1, and PB2, with a luciferase and β-galactosidase reporter. PA plasmids with the indicated PA residue 38 substitution were also transfected. Cells were either mock-treated (vehicle, DMSO, solid bars) or BXA-treated (10 nM, hashed bars) 1 h before transfection and 24 h or 48 h posttransfection. The polymerase activity luciferase output was normalized to β-galactosidase activity for each data point, then compared to the activity of the mock-treated I38-WT (100% activity, dotted line). Endonuclease data (in A and B) are presented as duplicate measures ± SD for each time point and concentration and are representative of at least three independent experiments. Minireplicon data (in C–F) are presented as at least duplicate measures ± SD and are representative of at least three independent assays for each virus polymerase complex. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001. Statistical comparison bar endcaps indicate differences from I38-WT mock-treated activity vs. an individual bar (┐) or both bars of the 38X substitution group (┤). Lack of comparison bars and P values on each graph represents “not significant.”