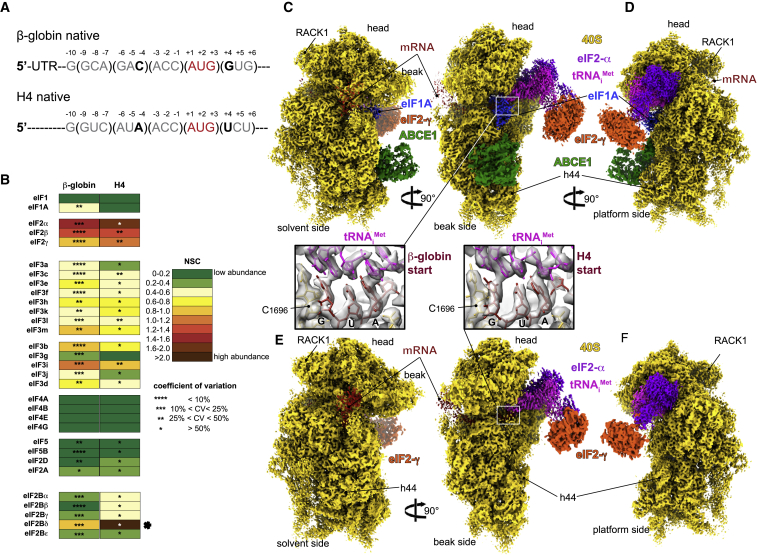
Figure 1.
Overall Structure of β-Globin and H4 Late-Stage 48S Initiation Complexes
(A) mRNA sequences used to form and purify the β-globin and the H4 ICs. Only the sequences near the AUG codon are represented, and main differences in the Kozak sequence are indicated in bold.
(B) Semiquantitative mass spectrometry analysis of the eIFs in both ICs, indicating the abundance of each eIF on the basis of the spectra count normalized. The two rounds of normalization were carried out using the total number of eIFs and estimated number of trypsin cleavage sites (see STAR Methods). The normalized spectra counts (NSCs) are presented as heatmaps, with cold colors indicating low abundance and warm colors indicating high abundance. The higher abundance of eIF2 proteins might be due to the excess of a free TC in the sample. The black star points out a large number of NSCs for eIF2Bδ, which is caused by the detection of three different isoforms of this protein. Small stars indicate the values of the coefficients of variation calculated for each NSC. In the analysis, the NSC for ABCE1 is not included, as it is a factor present also in other stages of translation than initiation.
(C and D) Segmented cryo-EM reconstructions of the β-globin IC seen from (C) solvent, beak and (D) platform sides, respectively. The reconstruction shows 40S (yellow), eIF2γ (orange), eIF2α (purple), tRNAiMet (magenta), mRNA (red), eIF1A (sky blue), and ABCE1 (green).
(E and F) Same as (C) and (D) but for the H4 IC. Boxed blowups represent the codon:anticodon duplexes in all shown reconstructions with their respective atomic models fitted in the corresponding electron densities.