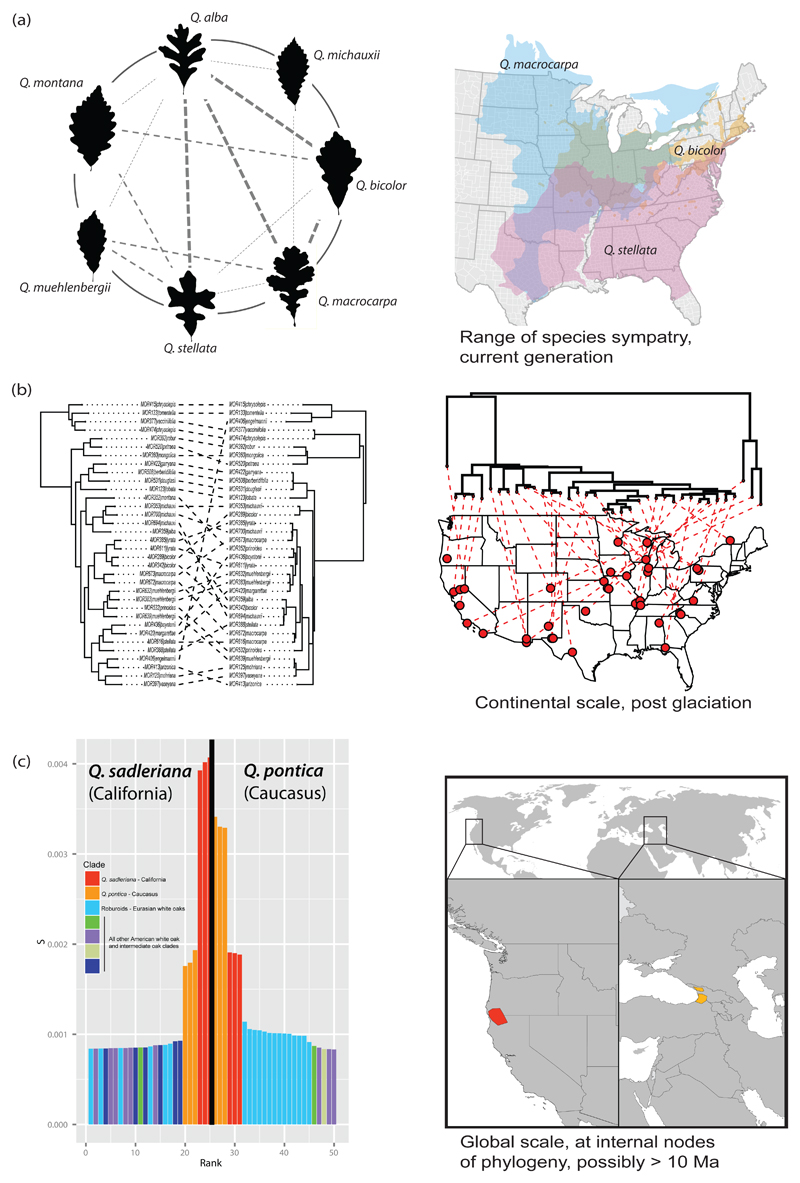
Figure 6. Contemporary introgression becomes ancient introgression.
(a) Contemporary introgression among the eastern North American white oaks has been a case study in the oak syngameon since the mid-1970s (Burger, 1975; Hardin, 1975; Van Valen, 1976) and was the focus of the first chloroplast phylogenetic study in oaks (Whittemore & Schaal, 1991). Introgression at this scale occurs primarily within the range of species sympatry and reflects recent generations of gene flow. The figure illustrates observed gene flow between seven eastern North American white oaks based on analysis of 80 SNPs that map back to all 12 Quercus linkage groups, with distances between uniquely mapped loci an average of 7.47 million bp (± 8.74 million bp, s.d.). The figure replicates the 16-species figure of Hardin 1975 (his Fig. 1), including only the subset of species investigated in Hipp et al. (2019b), with lines indicating hybridizations that Hardin inferred from morphological study. Thin dashed lines indicate hybridizations identified by Hardin but not by the SNP genotyping; medium dashed lines were identified by both Hardin and SNP genotyping, at an admixture level of 0.10 to 0.19 for at least one specimen; and thick dashed lines indicate SNP admixture levels of 0.20 or higher for at least one specimen. (Hipp et al. 2019b). (b) As species ranges change in response to climatic transitions, species that were once sympatric may become allopatric. Gene flow between them may still be detected by, for example, incongruence between chloroplast and nuclear topologies (Whittemore & Schaal, 1991; Manos et al., 1999). Here, a RAD-seq (nuclear) phylogeny for predominantly North American white oaks (left side) shows strong species cohesion, whereas the phylogeny of whole chloroplasts for the same samples sorts better by geography than by species (Pham et al., 2017). Such events may also be detected by phylogenomic incongruence in nuclear loci (McVay et al., 2017a; e.g., Kim et al., 2018). (c) With enough generations, ancient introgression events that were sufficiently pervasive may remain detectable. A fascinating case involves the sister species Q. pontica of the Caucasus, Q. sadleriana of California, and the Eurasian white oaks, or Roburoids. Because Q. pontica and Q. sadleriana are sister species, they are perforce equally closely related to all other oaks. With careful study, John McVay (2017b) and colleagues found that there are loci shared among the Roburoid white oaks that ally them much more closely to the Eurasian Q. pontica than to the American Q. sadleriana. Panel (c) shows genomic evidence for introgressive hybridization between wild Q. pontica samples and the Roburoid oaks: scaled locus similarity on the y-axis between Q. pontica and the Roburoids is significantly stronger than expected when data are simulated under incomplete lineage sorting alone (p < 0.002). Observed rankings of Roburoids to Q. sadleriana sample did not differ significantly from simulated distribution. The fact that these loci are shared among the Roburoids in spite of the fact that most are not sympatric with Q. pontica suggests a history of ancient introgression between the lineage that contains Q. pontica but not Q. sadleriana (allowing for extinction, there may have been other species) and an ancestral Roburoid. This finding is borne out with analysis of a separate phylogenomic dataset using species tree methods that account for hybridization. Thus a microevolutionary process, inferred to have preceded the crown divergence of the Roburoids an estimated 7.6-12.1 Ma (Hipp et al. 2019a), became frozen in the genome, like a fly in amber.