ABSTRACT
Feline infectious peritonitis virus (FIPV) may cause a lethal infection in cats. Antibody‐dependent enhancement (ADE) of FIPV infection has been recognized, and cellular immunity is considered to play an important role in preventing the onset of feline infectious peritonitis. In the present study, whether or not the T helper (Th)1 epitope was present in the spike (S)2 domain was investigated, the ADE epitope being thought to be absent from this domain. Three kinds of protein derived from the C‐terminal S2 domain of S protein of the FIPV KU‐2 strain were developed using a baculovirus expression system. These expressed proteins were the pre‐coil region which is the N‐terminal side of the putative fusion protein (FP), the region from FP to the heptad repeat (HR)2 (FP‐HR2) region, and the inter‐helical region which is sandwiched between HR1 and HR2. The ability of three baculovirus‐expressed proteins to induce Th1‐ and Th2‐type immune responses was investigated in a mouse model. It was shown that FP‐HR2 protein induced marked Th1‐ and Th2‐type immune responses. Furthermore, 30 peptides derived from the FP‐HR2 region were synthesized. Five and 16 peptides which included the Th1 and Th2 epitopes, respectively, were identified. Of these, four peptides which included both Th1 and Th2 epitopes were identified. These findings suggest that the identification of Th1 epitopes in the S2 domain of FIPV has important implications in the cat.
Keywords: coronavirus, feline infectious peritonitis virus, Th1‐type immune response
List of Abbreviations:
- ADE
antibody‐dependent enhancement
- CTL
cytotoxic T lymphocyte
- FIP
feline infectious peritonitis
- FIPV
feline infectious peritonitis virus
- FP
fusion protein
- HLA
human leukocyte antigen
- HR
heptad repeat
- IFN
interferon
- IH
inter‐helical
- IL
interleukin
- M
membrane
- MHC
major histocompatibility complex
- N
nucleocapsid
- PC
pre‐coil
- RT
reverse transcription
- S
spike
- SARS‐CoV
severe acute respiratory syndrome‐associated coronavirus
- SPF
specific‐pathogen‐free
- Tc
T cytotoxic
- TCID50
median tissue culture infective dose
- Th
T helper
FIP is a virus‐induced, chronically progressive, immunologically‐mediated, and usually fatal disease in domestic and wild Felidae. The causative agent of this disease is FIPV, which belongs to the family Coronaviridae, genus Coronavirus. In an attempt to prevent FIP, various vaccines, such as virulence‐attenuated live or inactivated FIPV vaccines, have been investigated. However none have shown sufficient efficacy, rather these vaccines have enhanced the onset of FIP (1, 2, 3, 4, 5, 6, 7). Compared to antibody‐negative kittens, intraperitoneal inoculation with FIPV induces more severe clinical signs in anti‐FIPV antibody‐positive kittens, and in kittens which have undergone passive immunization with serum or purified IgG from antibody‐positive cats (8, 9). This phenomenon is referred to as ADE of viral infection. The ADE of FIPV infection is a serious obstacle to its vaccine‐based prevention. Cellular immunity is considered to play an important role in preventing the onset of FIP (2). Thus, it is essential that vaccines against FIPV infection induce a cellular immune response.
FIPV consists of three major proteins, N, M, and S, which are classified as class I virus fusion proteins (10, 11). S protein exists as radially protruding trimers on the viral envelope, and can be structurally or functionally divided into two domains, namely the S1 and S2 domains, representing the N‐terminal globular head and C‐terminal membrane‐bound stalk, respectively. The C‐terminal S2 domain sequentially contains the PC region, putative FP, 4,3 hydrophobic HR1, IH domain, HR2, and the cluster aromatic amino acid domain from the N‐terminal, and is responsible for driving viral and target cell membrane fusion (10, 12). The N‐terminal S1 domain contains receptor‐binding, neutralizing antibody‐binding, and ADE epitopes (13, 14, 15, 16, 17, 18, 19). Recently, it was reported that Th1/Tc1 epitopes are present in the S1 and S2 domains of SARS‐CoV, which belongs to the Coronaviridae, as does FIPV (20, 21, 22, 23, 24, 25). However, it is not yet known whether the Th1/Tc1 epitope is present in the S1 and/or S2 domain of FIPV.
In the present study, we examined whether the Th1 epitope was present in the S2 domain, a domain in which it was thought that the ADE epitope was absent (13, 14, 16, 18, 19). We performed two experiments using a mouse model. Firstly, we investigated the immunogenicity of three baculovirus‐expressed proteins, PC, IH, and FP‐HR2 proteins, derived from the S2 domain of the type I FIPV KU‐2 strain (Fig. 1). It has been suggested that FP‐HR2 protein has marked immunogenicity. Secondly, we searched for epitopes inducing a Th1‐ and/or Th2‐type immune response using 30 synthetic peptides derived from the FP‐HR2 region. As a result, we identified 5 and 16 peptides including Th1 and Th2 epitopes, respectively.
Figure 1.
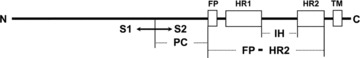
Schematic diagram of coronavirus S protein and the expressed regions of PC, FP‐HR2, and IH proteins. S1, the N‐terminal globular head of S protein; S2, the C‐terminal membrane‐bound stalk of S protein; TM, transmembrane.
MATERIALS AND METHODS
Preparation of recombinant baculovirus‐expressing proteins
Genomic RNA was extracted from FIPV KU‐2 strain‐infected culture fluid. RT of genomic RNA and amplification of cDNA employing PCR were carried out using a standard technique with a DNA thermal cycler. RT‐PCR primers were designed for each region: PC region (forward, 5′‐GGGGGATCCTTTTGTAACATCCTATACTATGCC‐3′; reverse, 5′‐GGGGAATTCTTAAGTGTACATAGACATCTTATTACCA‐3′), IH region (forward, 5′‐GGGAGATCTATGGCCCAAGTTGACCGT‐3′; reverse, 5′‐GGGGAATTCTTACACAATCTCCTGAAATGTC‐3′), and FP‐HR2 region (forward, 5′‐GGGAGATCTATGGCATCTTTAATTGGCGG‐3′; reverse, 5′‐GGGGAATTCTTATTTTACATAAGTTTCAATCCTGTT‐3′), of the S gene of the FIPV KU‐2 strain. The PCR products were cloned into a pVL 1392 plasmid (Pharmingen, San Diego, CA, USA). Escherichia coli TOP10 strain was transformed with recombinant plasmid DNA and cultured. The white colonies that grew were selected and recombinant bacmid DNA recovered. SF‐9 cells were transfected with recombinant bacmid DNA using cell Cellfectin (Gibco BRL, Grand Island, NY, USA), and the culture supernatant used in further experiments as each protein recombinant baculovirus (Fig. 1). Gene transfer was confirmed employing the PCR method (data not shown). SF‐9 cells cultured for a day were inoculated with the recombinant baculovirus. The infected cells were harvested and washed with PBS. One milliliter of RSB buffer (0.01 M NaCl, 0.0015 M MgCl2, and 0.01 M Tris‐HCl, pH 7.4) containing 0.2% NP‐40 was added to 1 × 107 cells, and the cell suspension kept at 4°C for 15 min with gentle shaking. The suspensions were centrifuged at 800 ×g for 10 min and the precipitate washed in PBS. The precipitate was then resuspended in PBS and used for immunization. The specificity and amount of each expressed protein were measured by Western immunoblotting using serum from the FIPV KU‐2 strain‐infected cat. Wild‐type baculovirus‐infected SF‐9 cells were prepared by NP‐40 treatment as control antigens for each expressed protein.
Western immunoblotting assay
Each expressed protein was separated employing 12% SDS‐PAGE and transferred to a nitrocellulose membrane. A standard protein marker (Precision Plus Protein Standards) was purchased from Bio‐Rad (Hercules, CA, USA). The membrane was blocked with 5% non‐fat dry milk powder in TBST (20 mM Tris‐HCl, pH 8.0, 0.88% NaCl, and 0.05% Tween‐20) for 1 hr at 37°C, incubated for 1 hr at 37°C with serum from the FIPV KU‐2 strain‐infected cat and then incubated with peroxidase‐conjugated goat anti‐cat IgG (MP Biomedicals, LLC‐Cappel products, Irvine, CA, USA) for 1 hr at 37°C. It was then visualized in substrate for 10 min.
Peptide synthesis
To determine the Th1 and/or Th2 epitope, 30 peptides derived from the S2 domain of the FIPV KU‐2 strain were synthesized at Sigma‐Aldrich (St Louis, MO, USA) (Table 1). One peptide (FP) was derived from the putative FP region. Twenty‐five peptides (HR1‐1–HR1‐14 and HR2‐1–HR2‐11) synthesized as 20‐mer fragments with a 12‐amino‐acid overlap were derived from the HR1 and HR2 regions. Four peptides (IH‐1–IH‐4) were derived from the hydrophobic area of the IH domain. All peptides were purified to purities of more than 70% and supplied as lyophilized powder. The peptides were dissolved in 10% dimethyl sulfoxide at 1 mg/ml, aliquoted, and stored at −80°C.
Table 1.
Amino acid sequences of the peptides derived from the S2 domain of the FIPV KU‐2 strain
| Peptide name | Amino acid sequence | Start position |
|---|---|---|
| FP | GNKMSMYTASL | 1029 |
| HR1‐1 | TSAVAVPFAMQVQARLNYVA | 1049 |
| HR1‐2 | AMQVQARLNYVALQTDVLQE | 1057 |
| HR1‐3 | NYVALQTDVLQENQKILANA | 1065 |
| HR1‐4 | VLQENQKILANAFNNAIGNI | 1073 |
| HR1‐5 | LANAFNNAIGNITLALGKVS | 1081 |
| HR1‐6 | IGNITLALGKVSNAITTTSD | 1089 |
| HR1‐7 | GKVSNAITTTSDGFNSMASA | 1097 |
| HR1‐8 | TTSDGFNSMASALTKIQSVV | 1105 |
| HR1‐9 | MASALTKIQSVVNQQGEALS | 1113 |
| HR1‐10 | QSVVNQQGEALSQLTSQLQK | 1121 |
| HR1‐11 | EALSQLTSQLQKNFQAISSS | 1129 |
| HR1‐12 | QLQKNFQAISSSIAEIYNRL | 1137 |
| HR1‐13 | ISSSIAEIYNRLEKVEADAQ | 1145 |
| HR1‐14 | YNRLEKVEADAQVDRLITGR | 1153 |
| IH‐1 | ITGRLAALNAYVSQTLTQY | 1169 |
| IH‐2 | FSLVNSAPEGLLFFHTVLLPTEWEEVTA | 1222 |
| IH‐3 | FFHTVLLPTEWEEVTAWSGIC | 1234 |
| IH‐4 | WSGICVNDTYAYVLKDFDHSIFSYNGTY | 1250 |
| HR2‐1 | TFQEIVIDYIDINKTIADML | 1312 |
| HR2‐2 | YIDINKTIADMLEQYNPNYT | 1320 |
| HR2‐3 | ADMLEQYNPNYTTPELNLLL | 1328 |
| HR2‐4 | PNYTTPELNLLLDIFNQTKL | 1336 |
| HR2‐5 | NLLLDIFNQTKLNLTAEIDQ | 1344 |
| HR2‐6 | QTKLNLTAEIDQLEQRADNL | 1352 |
| HR2‐7 | EIDQLEQRADNLTTIAHELQ | 1360 |
| HR2‐8 | ADNLTTIAHELQQYIDNLNK | 1368 |
| HR2‐9 | HELQQYIDNLNKTLVDLDWL | 1376 |
| HR2‐10 | NLNKTLVDLDWLNRIETYVK | 1384 |
| HR2‐11 | LDWLNRIETYVKWPWYVWLL | 1392 |
Immunization of mice
Four‐week‐old female BALB/c mice (n= 3 or 4 per group) were obtained from Charles River (Hino, Japan) and maintained under SPF conditions. All immunizations were given intraperitoneally every two weeks with each expressed protein (Western immunoblotting: 16 units per dose). Mice were immunized three times in total. Wild‐type baculovirus‐infected SF‐9 cell‐derived antigen and PBS‐immunized groups were used as negative controls.
Stimulation of splenocytes
One or two weeks after the final immunization, the mice were killed to harvest their spleens. Splenocytes were cultured in RPMI‐1640 medium containing 5% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin. Erythrocyte‐depleted splenocytes (4 × 105 cells/well) in 96‐well plates were cultured with 30 μg/mL of each peptide, heat‐inactivated virus (FIPV KU‐2 strain, 104.6 TCID50/mL; FIPV 79–1146 strain, 105.5 TCID50/mL; the homology of the amino acid sequence of the S2 domain of these two viruses is 60.7%), or culture medium as a control for three days.
ELISA
The concentrations of IFN‐γ, IL‐2, and IL‐4 in the supernatant of splenocytes cultured with stimulatory substances for three days were measured using commercial mouse IFN‐γ, IL‐2, and IL‐4 ELISA kits (Endogen, Cambridge, MA, USA) according to the manufacturer's protocol, respectively.
Statistical analysis
All results are expressed as means ± standard error of the mean. Student's t‐test or one‐way analysis of variance was employed as appropriate to determine the significance of differences. A P‐value of < 0.01 or P < 0.05 was considered significant.
RESULTS
Specificity and amount of each expressed protein
The specificity and amount of three expressed proteins, PC, FP‐HR2, and IH, were measured by Western immunoblotting assay using serum from the FIPV KU‐2 strain‐infected cat. In agreement with the size of PC, FP‐HR2, and IH proteins, 25‐, 37‐, and 15‐kDa bands, respectively, were visualized (Fig. 2) and detected in up to a 16‐fold dilution (16 units). No expressed proteins reacted with the serum of the SPF cat.
Figure 2.
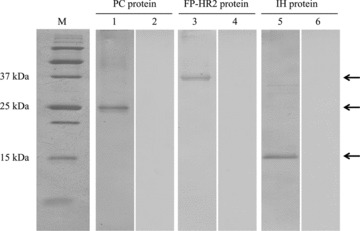
Analysis of expressed proteins using Western immunoblotting. M, marker: lanes 1 and 2, PC protein: lanes 3 and 4, FP‐HR2 protein: lanes 5 and 6, IH protein. Lanes 1, 3, and 5, serum of the FIPV KU‐2 strain‐infected cat; lanes 2, 4, and 6, serum of the SPF cat. The arrows show expressed proteins, PC, FP‐HR2, and IH proteins.
Th1‐ and Th2‐type immune responses against heat‐inactivated FIPV antigen of murine splenocytes immunized with each expressed protein
Evaluation of the Th1‐type immune response of PC, FP‐HR2, and IH proteins was based on the concentrations of IFN‐γ and IL‐2 in the culture supernatant (Fig. 3a, b). In FP‐HR2 protein‐immunized mice, the concentrations of IFN‐γ and IL‐2 in the supernatant of splenocytes cultured with or without FIPV antigen were significantly higher than in that of splenocytes derived from wild‐type baculovirus‐infected SF‐9‐ or PBS‐immunized mice (P < 0.05, P < 0.01, respectively). In IH protein‐immunized mice, the concentration of IL‐2 in the supernatant of splenocytes cultured with or without FIPV antigen was significantly higher than in that of splenocytes derived from wild‐type baculovirus‐infected SF‐9‐ or PBS‐immunized mice (P < 0.05, P < 0.01, respectively). However, in PC protein‐immunized mice, the concentrations of IFN‐γ and IL‐2 in the supernatant of splenocytes cultured with or without FIPV antigen were not significantly higher than in that of splenocytes derived from wild‐type baculovirus‐infected SF‐9‐ or PBS‐immunized mice.
Figure 3.
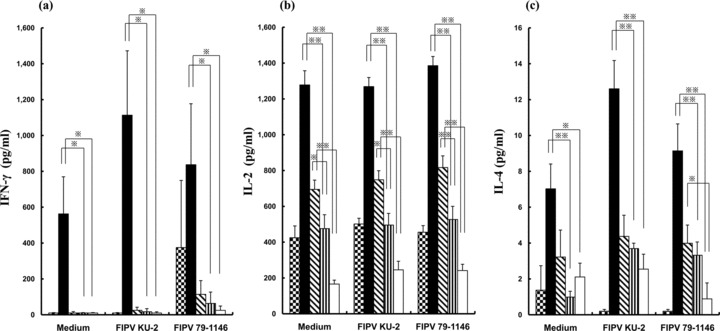
Splenocytes derived from mice immunized with PC protein, FP‐HR2 protein, IH protein, Wild‐type baculovirus‐infected SF‐9 or PBS were cultured with heat‐inactivated FIPV (KU‐2 or 79–1146 strain) or culture medium as a negative control for three days. The concentrations of (a) IFN‐γ, (b) IL‐2 and (c) IL‐4 in the culture medium were measured using an ELISA kit. *, P < 0.05; *, P < 0.01 compared with the wild‐type baculovirus‐infected SF‐9‐ or PBS‐immunized group;  , mice immunized with PC protein; (▪), mice immunized with FP‐HR2 protein;
, mice immunized with PC protein; (▪), mice immunized with FP‐HR2 protein;  , mice immunized with IH protein;
, mice immunized with IH protein;  , mice immunized with Wild‐type baculovirus‐infected SF‐9; (□), mice immunized with PBS.
, mice immunized with Wild‐type baculovirus‐infected SF‐9; (□), mice immunized with PBS.
Evaluation of the Th2‐type immune response of PC, FP‐HR2, and IH proteins was based on the concentration of IL‐4 in the supernatant (Fig. 3c). In FP‐HR2 protein‐immunized mice, the concentration of IL‐4 in the supernatant of splenocytes cultured with or without FIPV antigen was significantly higher than in that of splenocytes derived from wild‐type baculovirus‐infected SF‐9‐ or PBS‐immunized mice (P < 0.05, P < 0.01). In IH protein‐immunized mice, the concentration of IL‐4 in the supernatant of splenocytes cultured with the heat‐inactivated FIPV 79–1146 strain was significantly higher than in that of splenocytes derived from PBS‐immunized mice (P < 0.05). However, in PC protein‐immunized mice, the concentration of IL‐4 in the supernatant of splenocytes cultured with or without FIPV antigen was not significantly higher than in that of splenocytes derived from wild‐type baculovirus‐infected SF‐9‐ or PBS‐immunized mice.
Th1‐ and Th2‐type immune responses against peptides derived from the FP‐HR2 region of murine splenocytes immunized with FP‐HR2 protein
Evaluation of the Th1‐ or Th2‐type immune response against 30 peptides derived from the FP‐HR2 region was based on the concentration of IFN‐γ or IL‐4 in the supernatant, respectively (4, 5). In FP‐HR2 protein‐immunized mice, the concentration of IFN‐γ in the supernatant of splenocytes cultured with the HR1‐7, HR1‐14, IH‐2, HR2‐4, HR2‐11, or FIPV antigen was significantly higher than in that of splenocytes cultured with medium alone (P < 0.01: IH‐2, HR2‐4, HR2‐11, and FIPV antigen; P < 0.05: HR1‐7 and HR1‐14) (Fig. 4a). The concentration of IL‐4 in the supernatant of splenocytes cultured with the FP, HR1‐6, HR1‐7, HR1‐10, HR1‐11, HR1‐12, HR1‐14, IH‐1, IH‐2, IH‐4, HR2‐1, HR2‐2, HR2‐3, HR2‐7, HR2‐8, HR2‐11, or FIPV antigen was significantly higher than in that of splenocytes cultured with medium alone (P < 0.01: HR1‐7, HR1‐10, HR1‐11, HR1‐12, HR1‐14, HR2‐2, HR2‐3, HR2‐7, HR2‐8, HR2‐11, and FIPV antigen; P < 0.05: FP, HR1‐6, IH‐1, IH‐2, IH‐4, and HR2‐1) (Fig. 5a). This indicates that four peptides, HR1‐7, HR1‐14, IH‐2, and HR2‐11, induce both Th1‐ and Th2‐type immune responses. In wild‐type baculovirus‐infected SF‐9‐ or PBS‐immunized mice, the concentrations of IFN‐γ and IL‐4 in the supernatant of splenocytes cultured with or without peptide were not increased (Fig. 4b, c; Fig. 5b, c).
Figure 4.
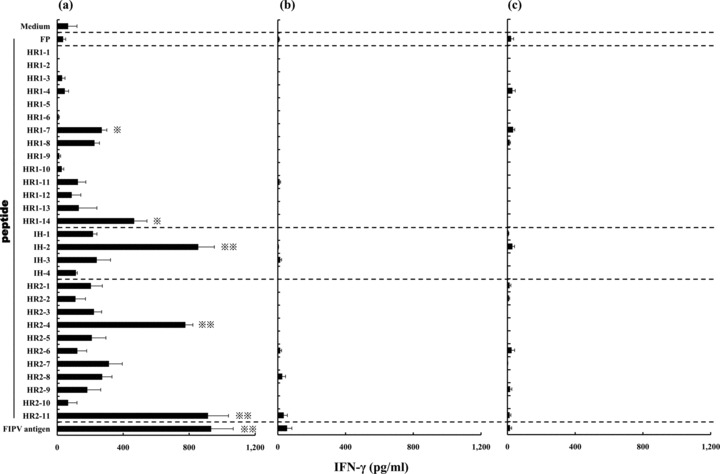
Splenocytes derived from mice immunized with (a) FP‐HR2 protein, (b) wild‐type baculovirus‐infected SF‐9 or (c) PBS were cultured with individual peptides derived from the FP‐HR2 region of the FIPV KU‐2 strain, heat‐inactivated FIPV KU‐2 strain as a positive control, or culture medium as a negative control for three days. The concentration of IFN‐γ in the supernatant was measured using a Mouse ELISA kit. *, P < 0.05; **, P < 0.01 compared with the control culture with medium alone.
Figure 5.
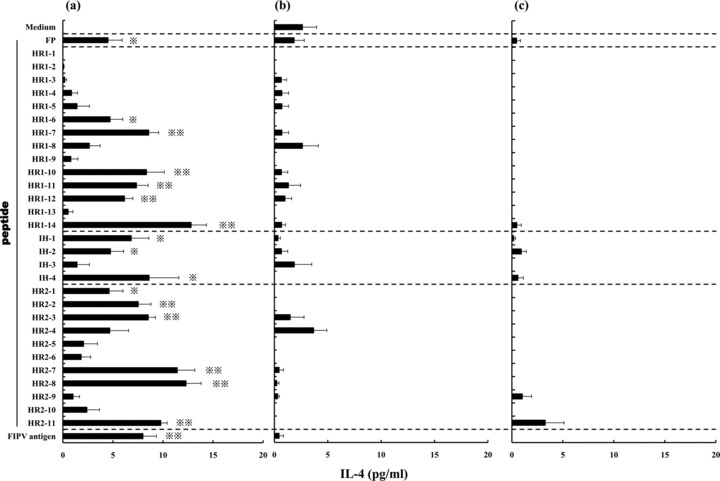
Splenocytes derived from mice which were immunized with (a) FP‐HR2 protein, (b) wild‐type baculovirus‐infected SF‐9 or (c) PBS were cultured with individual peptides derived from the FP‐HR2 region of the FIPV KU‐2 strain, heat‐inactivated FIPV KU‐2 strain as a positive control, or culture medium as a negative control for three days. The concentration of IL‐4 in the supernatant was measured using a mouse ELISA kit. *, P < 0.05; **, P < 0.01 compared with the control culture with medium alone.
DISCUSSION
We have shown that FP‐HR2 protein derived from the S2 domain, which does not include ADE epitopes, exhibits strong immunogenicity in the mouse. In addition, we showed that 5 and 15 peptides derived from the FP‐HR2 region induce Th1‐ and Th2‐type immune responses, respectively. These results suggest that these peptides contain the Th1 and Th2 epitopes, respectively. Of these, four peptides contained both the Th1 and Th2 epitopes.
Comparing the immunogenicity of mouse N‐terminal‐sided PC protein with that of C‐terminal‐sided FP‐HR2 protein of the S2 domain, IFN‐γ, IL‐2, and IL‐4 concentrations were significantly increased only in FP‐HR2 protein‐immunized mice. FP‐HR2 protein consists of FP, HR1, HR2, and IH domains. The concentrations of IFN‐γ, IL‐2, and IL‐4 in FP‐HR2 protein‐immunized mice were significantly higher than those of IH protein‐immunized mice. This suggests that, except for the IH domain, the FP, HR1, and/or HR2 region contains a highly immunogenic site. IH protein showed less immunogenicity in mice, especially in regard to induction of IFN‐γ, than did FP‐HR2 protein. However, stimulation with IH‐2, a peptide derived from the IH region, induced IFN‐γ production, suggesting the presence of Th1 epitopes. Reportedly, SARS‐CoV and transmissible gastroenteritis virus produce no neutralizing antibodies after immunization of mice with a peptide containing a neutralizing epitope, but enhance immunogenicity and produce neutralizing antibodies when bound to a peptide with Th1 epitopes (26, 27). The HR1 and HR2 regions contain peptides with the Th1 epitopes identified in this study, (i.e. HR1‐7, HR1‐14, HR2‐4, and HR2‐11). Thus, IH protein alone is unlikely to recognize antigen‐presenting cells; however, FP‐HR2 protein (i.e. IH protein bound to HR1 and HR2) may exhibit greater immunogenicity.
H‐2Db, an MHC class I molecule of BALB/c mice, is known to have a structure similar to that of HLA‐A24. Amino acid sequences predicted by the National Institute of Health (http://www-bimas.cit.nih.gov/molbio/hla_bind/) to have a strong capacity to bind to HLA‐A24, were explored in the FP, HR1, and HR2 regions. The results demonstrated the presence of epitopes showing a marked capacity to bind to aa1109‐1117 (HR1‐8) of the HR1 region and aa1319‐1327 (HR2‐1), aa1337‐1345 (HR2‐3 and HR2‐4), aa1349‐1357 (HR2‐5), and aa1406‐1414 (HR2‐11) of the HR2 region. Among these peptides, HR2‐4 and HR2‐11 induced IFN‐γ production in this experiment. This suggests that these two peptides are MHC class I‐binding peptides with the Tc1 epitope. The other peptides, HR1‐7, HR1‐14, and IH‐2, have the Th1 epitope.
Like the sites of Th1 epitopes in FIPV identified in this study, SARS‐CoV contains Th1/Tc1 epitopes in the HR1, IH, and HR2 regions of the S2 domain (22, 23, 24). Among these, Th1/Tc1 epitopes (SSp‐1) present in the HR2 region are known to induce strong CTL activity and are regarded as candidates for vaccine development. Thus, the peptides with Th1/Tc1 epitopes identified in this study, particularly HR2‐4 and HR2‐11 that seem to have Tc1 epitopes, are potential candidates for vaccine development for prevention of FIPV infection.
Although Th1/Tc1 epitopes in the FP‐HR2 region appeared to strongly induce the Th1‐type immune response in a mouse model, their effects in the cat remain unknown. It is important for an effective peptide‐based vaccine against FIPV infection to include Th1/Tc1, but not ADE, epitopes. In the cat, the identification of Th1/Tc1 epitopes in the S2 domain of FIPV has important implications. In the future, Th1/Tc1 epitopes in FIPV will be explored using FIPV‐infected cats. The development of a vaccine against FIPV infection based on these epitopes is awaited.
REFERENCES
- 1. Barlough J.E., Stoddart C.A., Sorresso G.P., Jacobson R.H., Scott F.W. (1984) Experimental inoculation of cats with canine coronavirus and subsequent challenge with feline infectious peritonitis virus. Lab Anim Sci 34: 592–7. [PubMed] [Google Scholar]
- 2. Pedersen N.C. (2009) A review of feline infectious peritonitis virus infection: 1963–2008. J Feline Med Surg 11: 225–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pedersen N.C., Black J.W. (1983) Attempted immunization of cats against feline infectious peritonitis, using avirulent live virus or sublethal amounts of virulent virus. Am J Vet Res 44: 229–34. [PubMed] [Google Scholar]
- 4. Pedersen N.C., Boyle J.F., Floyd K., Fudge A., Barker J. (1981) An enteric coronavirus infection of cats and its relationship to feline infectious peritonitis. Am J Vet Res 42: 368–77. [PubMed] [Google Scholar]
- 5. Pedersen N.C., Evermann J.F., Mckeirnan A.J., Ott R.L. (1984) Pathogenicity studies of feline coronavirus isolates 79–1146 and 79–1683. Am J Vet Res 45: 2580–5. [PubMed] [Google Scholar]
- 6. Stoddart C.A., Barlough J.E., Baldwin C.A., Scott F.W. (1988) Attempted immunisation of cats against feline infectious peritonitis using canine coronavirus. Res Vet Sci 45: 383–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Woods R.D., Pedersen N.C. (1979) Cross‐protection studies between feline infectious peritonitis and porcine transmissible gastroenteritis viruses. Vet Microbiol 4: 11–6. [Google Scholar]
- 8. Pedersen N.C., Boyle J.F. (1980) Immunologic phenomena in the effusive form of feline infectious peritonitis. Am J Vet Res 41: 868–76. [PubMed] [Google Scholar]
- 9. Weiss R.C., Scott F.W. (1981) Antibody‐mediated enhancement of disease in feline infectious peritonitis: comparisons with dengue hemorrhagic fever. Comp Immunol Microbiol Infect Dis 4: 175–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bosch B.J., Van Der Zee R., De Haan C.A., Rottire P.J. (2003) The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J Virol 77: 8801–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Olsen C.W. (1993) A review of feline infectious peritonitis virus: molecular biology, immunopathogenesis, clinical aspects, and vaccination. Vet Microbiol 36: 1–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dimitrov D.S. (2004) Virus entry: molecular mechanisms and biomedical applications. Nat Rev Microbiol 2: 109–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Corapi W.V., Darteil R.J., Audonnet J.C., Chappuis G.E. (1995) Localization of antigenic sites of the S glycoprotein of feline infectious peritonitis virus involved in neutralization and antibody‐dependent enhancement. J Virol 69: 2858–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Corapi W.V., Olsen C.W., Scott F.W. (1992) Monoclonal antibody analysis of neutralization and antibody‐dependent enhancement of feline infectious peritonitis virus. J Virol 66: 6695–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hohdatsu T., Nakamura M., Ishizuka Y., Yamada H., Koyama H. (1991) A study of the mechanism of antibody‐dependent enhancement of feline infectious peritonitis virus infection in feline macrophages by monoclonal antibodies. Arch Virol 120: 207–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hohdatsu T., Yamada H., Ishizuka Y., Koyama H. (1993) Enhancement and neutralization of feline infectious peritonitis virus infection in feline macrophages by neutralizing monoclonal antibodies recognizing different epitopes. Microbiol Immunol 37: 499–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hohdatsu T., Yamada M., Tominaga R., Makino K., Kida K., Koyama H. (1998) Antibody‐dependent enhancement of feline infectious peritonitis virus infection in feline alveolar macrophages and human monocyte cell line U937 by serum of cats experimentally or naturally infected with feline coronavirus. J Vet Med Sci 60: 49–55. [DOI] [PubMed] [Google Scholar]
- 18. Kida K., Hohdatsu T., Fujii K., Koyama H. (1999) Selection of antigenic variants of the S glycoprotein of feline infectious peritonitis virus and analysis of antigenic sites involved in neutralization. J Vet Med Sci 61: 935–8. [DOI] [PubMed] [Google Scholar]
- 19. Olsen C.W., Corapi W.V., Ngichabe C.K., Baines J.D., Scott F.W. (1992) Monoclonal antibodies to the spike protein of feline infectious peritonitis virus mediate antibody‐dependent enhancement of infection of feline macrophages. J Virol 66: 956–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bai B., Lu X., Meng J., Hu Q., Mao P., Lu B., Chen Z., Yuan Z., Wang H. (2008) Vaccination of mice with recombinant baculovirus expressing spike or nucleocapsid protein of SARS‐like coronavirus generates humoral and cellular immune responses. Mol Immunol 45: 868–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen H., Hou J., Jiang X., Ma S., Meng M., Wang B., Zhang M., Zhang M., Tang X., Zhang F., Wan T., Li N., Yu Y., Hu H., Yang R., He W., Wang X., Cao X. (2005) Response of memory CD8+ T cells to severe acute respiratory syndrome (SARS) coronavirus in recovered SARS patients and healthy individuals. J Immunol 175: 591–8. [DOI] [PubMed] [Google Scholar]
- 22. Tsao Y.P., Lin J.Y., Jan J.T., Leng C.H., Chu C.C., Yang Y.C., Chen S.L. (2006) HLA‐A*0201 T‐cell epitopes in severe acute respiratory syndrome (SARS) coronavirus nucleocapsid and spike proteins. Biochem Biophys Res Commun 344: 63–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang B., Chen H., Jiang X., Zhang M., Wan T., Li N., Zhou X., Wu Y., Yang F., Yu Y., Wang X., Yang R., Cao X. (2004) Identification of an HLA‐A*0201‐restricted CD8+ T‐cell epitope SSp‐1 of SARS‐CoV spike protein. Blood 104: 200–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang Y.D., Sin W.Y., Xu G.B., Yang H.H., Wong T.Y., Pang X.W., He X.Y., Zhang H.G., Ng J.N., Cheng C.S., Ju J., Meng L., Yang R.F., Lai S.T., Guo Z.H., Xie Y., Chen W.F. (2004) T‐cell epitopes in severe acute respiratory syndrome (SARS) coronavirus spike protein elicit a specific T‐cell immune response in patients who recover from SARS. J Virol 78: 5612–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhou M., Xu D., Li X., Li H., Shan M., Tang J., Wang M., Wang F.S., Zhu X., Tao H., He W., Tien P., Gao G.F. (2006) Screening and identification of severe acute respiratory syndrome‐associated coronavirus‐specific CTL epitopes. J Immunol 177: 2138–45. [DOI] [PubMed] [Google Scholar]
- 26. Antón I.M., Suñé C., Meloen R.H., Borrás‐Cuesta F., Enjuanes L. (1995) A transmissible gastroenteritis coronavirus nucleoprotein epitope elicits T helper cells that collaborate in the in vitro antibody synthesis to the three major structural viral proteins. Virology 212: 746–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhao J., Huang Q., Wang W., Zhang Y., Lv P., Gao X.M. (2007) Identification and characterization of dominant helper T‐cell epitopes in the nucleocapsid protein of severe acute respiratory syndrome coronavirus. J Virol 81: 6079–88. [DOI] [PMC free article] [PubMed] [Google Scholar]


