ABSTRACT
Unlike for serotype II feline coronaviruses (FCoV II), the cellular receptor for serotype I FCoV (FCoV I), the most prevalent FCoV serotype, is unknown. To provide a platform for assessing the pattern by which FCoV I attaches to its host receptor(s), HEK293 cell lines that stably express the ectodomains of the spike (S) proteins derived from a FCoV I feline enteric coronavirus strain UU7 (FECV UU7) and a feline infectious peritonitis virus strain UU4 (FIPV UU4) were established. Using the recombinant S proteins as probes to perform S protein affinity histochemistry in paraffin‐embedded tissues, although no tissue or enteric binding of FECV UU7 S protein was detected, it was found that by immunohistochemistry that the tissue distribution of FIPV UU4 S protein‐bound cells correlated with that of FIPV antigen‐positive cells and lesions associated with FIP and that the affinity binding of FIPV UU4 S protein on macrophages was not affected by enzymatic removal of host cell‐surface sialic acid with neuraminidase. These findings suggest that a factor(s) other than sialic acid contribute(s) to the macrophage tropism of FIPV strain UU4. This approach allowed obtaining more information about both virus–host cell interactions and the biological characteristics of the unidentified cellular receptor for FCoV I.
Keywords: cell tropism, feline infectious peritonitis virus, histochemistry, spike
List of Abbreviations
- APN
aminopeptidase N
- CRFK
Crandell Rees feline kidney
- FCoV
feline coronaviruses
- FCoVI
serotype I feline coronaviruses
- FCoVII
serotype II feline coronaviruses
- FCWF‐4 cells
Felis catus whole fetus‐4 cells
- FECV
feline enteric coronavirus
- FIP
feline infectious peritonitis
- FIPV
feline infectious peritonitis virus
- IHC
immunohistochemistry staining
- NA
neuraminidase
- PBMC
peripheral blood mononuclear cell
- PVDF
polyvinylidene difluoride
- RT
room temperature
- S
spike
- TBST
Tris‐buffered saline with 0.05% Tween 20
Coronaviruses (subfamily Coronavirinae, order Nidovirales) are enveloped, positive‐stranded RNA viruses that exhibit a broad host range and cause respiratory and/or intestinal infections in various avian and mammalian species 1. The emergence of severe acute respiratory syndrome coronavirus in 2002 and Middle East respiratory syndrome coronavirus in 2012 demonstrated that coronaviruses have the potential to switch hosts 2, 3. The evolution and emergence of coronaviruses are attributable to mutations and recombination between their large RNA genomes 4, 5. Binding of virus to host cells plays a critical role in determining tissue tropism, host specificity and pathogenicity of that virus. The S protein of coronaviruses plays crucial roles in binding and fusion to host cells 6.
Feline coronavirus belongs to the genus Alphacoronavirus and consists of two serotypes that are defined by serological and genetic characteristics of the S protein 7, 8, 9. Although highly prevalent 10, 11, FCoV I are less well understood than FCoV II because of the former's poor growth in cell culture. In contrast, serotype II FCoVs are less prevalent but have been studied in a relatively detailed manner because they are easily propagated in vitro. Feline APN is reportedly the functional receptor for FCoV II 12, whereas sialic acid is considered a co‐receptor for FCoV enterotropism 13. However, the primary cellular receptor for FCoV I has not yet been identified 14.
Each serotype of FCoV comprises two biotypes: the avirulent FECV and the lethal FIPV. The prevalence of FECV in cats living in multi‐cat households reaches 90% and this biotype is associated with asymptomatic to mild, self‐limiting gastrointestinal disease 15, 16. In contrast, FIPVs show a tropism for macrophages 17, 18, 19. Some investigators have suggested that FIPVs and FECVs are independently circulating viruses 20, 21. However, accumulating evidence has shown that FIPVs arise from genetic mutations of FECVs during in vivo infection 15, 22, 23, 24, 25, 26, 27, 28, leading to switching of tropism from intestinal enterocytes to motile monocytes/macrophages and causing systemic dissemination of the virus and FIP. FIP, a fatal immunopathologic disease, is characterized by disseminated pyogranulomas and severe inflammatory damage around blood vessels 26, 28. Mutations in the S protein of FCoVs appear to be responsible for the change in viral tropism 5, 29.
Using a viral protein as a probe to study the interactions of a virus with its host cellular receptor in paraffin‐embedded tissues is known as virus protein histochemistry. Several studies have used this approach to determine the distribution of virus receptors and their association with the virus's cell tropism 30, 31. To date, the cellular receptor for FCoV I is unknown; hence, it is difficult to propagate the virus in cell culture for further study. To gain information on the cell tropism of this virus and to provide a platform for assessing S–host receptor binding, we constructed HEK293 cell lines that stably express the ectodomain of S proteins derived from the FIPV strain UU4 and the FECV strain UU7. Using recombinant S protein as a probe, we performed S protein histochemistry on paraffin‐embedded tissues from cats confirmed to have FIP infection and cats without FIP infection, with or without enzymatic removal of cell‐surface sialic acid with NA. Our results show that binding of the FIPV S protein correlates with the cell tropism of the virus and reflects FIP pathogenesis.
MATERIALS AND METHODS
Construction and expression of the ectodomain of FIPV strain UU4 and FECV strain UU7 S protein
The nucleotide sequence spanning the ectodomain (amino acid positions 1 to 1408) of S sequences of FIPV strain UU4 (GenBank accession number FJ938054) and FECV strain UU7 (GenBank accession number FJ938053) were codon optimized, after which synthetic genes were produced by Genscript (Piscataway, NJ, USA). Each synthetic gene was subcloned into BamHI‐NotI restriction sites of the pcDNA 3.1‐V5‐His vector (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions (Fig. 1a). Plasmid DNA was purified using a QIAprep Spin Miniprep Kit (Qiagen, Hilden, Germany). For protein expression, HEK293 cells were transfected with each plasmid for 48 hr using PolyJet transfection reagent (SignaGen, Gaithersburg, MD, USA) according to the manufacturer's instructions. At 48 hr post‐transfection, the plasmid‐transfected cells were selected by replacing the medium with selection medium, consisting of complete Dulbecco's modified Eagle's medium (Gibco, Gaithersburg, MD, USA) supplemented with 10% FBS and 750 µg/mL Geneticin (Invitrogen) for two weeks to establish a cell line that stably expresses FIPV UU4 S protein (HEK‐UU4 S cells) and FECV UU7 S protein (HEK‐UU7 S cells). Expressions of S proteins in the HEK‐UU4S and HEK‐UU7S cell lines were detected using immunocytochemical staining and western blotting using an anti‐V5 antibody (Invitrogen).
Figure 1.
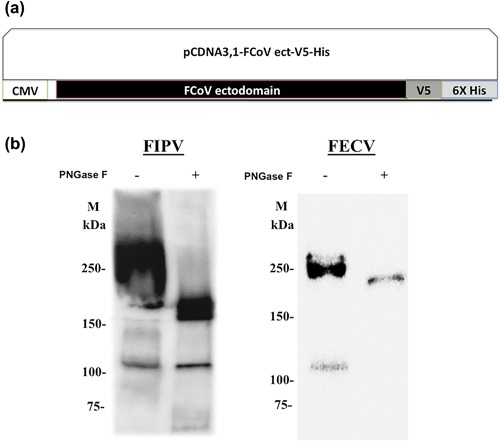
Expression of FCoV I S ectodomain. (a) Schematic representation of the sequence of the ectodomain of S protein derived from serotype I FIPV UU4 strain or FECV UU7 strain cloned in frame in a pcDNA 3.1‐V5‐His vector under the control of a cytomegalovirus promoter. (b) Western blot detection of S protein ectodomain expression in HEK293 cells. The expressed FIPV UU4 S and FECV UU7 S proteins, with (PNGase +) or without (PNGase −) peptide N‐glycosidase treatment, were separated by SDS‐PAGE, followed by western blotting, and detected by anti‐V5 tag mAb and goat anti‐mouse antibody conjugated with HRP. CMV, cytomegalovirus; M: marker.
Protein expression and purification
HEK‐UU4S and HEK‐UU7S cells were grown in 150T flasks (Corning, Cambridge, MA, USA) in the selection medium and incubated at 37°C in 5% CO2 for 108 hr. Cell supernatants were collected by centrifugation for 10 min at 1000 rpm and filtered through a 0.22 μm pore size membrane. To prepare cell lysates, cells were washed with Dulbecco's PBS (Life Technologies, Grand island, NY, USA) twice and then incubated with RIPA cell lysis buffer (Amresco, Solon, OH, USA) at 4°C for 20 min. The lysates were clarified by centrifugation for 10 min at 1000 rpm and filtered through a 0.22 μm pore size membrane. In accordance with the manufacturer's protocol, a 50% slurry of resin (TALON Superflow; GE Healthcare, Little Chalfont, UK) was incubated with supernatants or cell lysate collected from HEK‐UU4S cells in binding buffer (50 mM Na2HPO4, 0.15 M NaCL, 10 mM imidazole, pH 8.0). Purified S protein was eluted, desalted and concentrated with a Vivaspin 20 MWCO 100 kDa (GE Healthcare) according to the manufacturer's instructions and stored at −80°C. The concentration of the purified protein was determined by using a Picodrop microliter spectrophotometer (Saffron Walden, UK) with an absorbance at 280 nm. PBS was used as a blank.
Western blotting
Expression and purification of S protein was confirmed by SDS‐PAGE followed by western blotting using an anti‐V5 antibody (Invitrogen). Briefly, S protein was diluted with distilled water, sample buffer (4× dilution), and dl‐dithiothreitol (10× dilution) to a volume of 40 µL (26 µL diluted S protein, 10 µL sample buffer, 4 µL dl‐dithiothreitol) and heated at 95°C for 10 min. After electrophoresis and transfer, PVDF membranes (Bio‐Rad, Hercules, CA, USA) were blocked with 5% skim milk (diluted with TBST) for 1 hr and washed with TBST. The PVDF membranes were first incubated with 5000× mouse anti‐V5 Tag antibody (Life Technologies, Carlsbad, CA, USA) for 1 hr and incubated with 10,000× HRP conjugated goat anti‐mouse antibody (Jackson ImmunoResearch Laboratories, West Grove, PA, USA) for 1 hr for detection. Binding was visualized using Clarity Western ECL Blotting Substrate (Bio‐Rad) according to the manufacturer's instructions.
Enzymatic removal of N‐glycans of the recombinant S proteins
The S proteins were digested by peptide N‐glycosidase F (PNGase F; New England Biolabs, Ipswich, MA, USA) according to the manufacturer's instructions. Briefly, 10–20 μg of S protein was mixed with 2 µL of glycoprotein denaturing buffer (10× dilution) and distilled deionized water added to a total volume of 10 µL. The S protein mixture was denatured by heating at 100°C for 10 min and immediately cooled down on ice. After the solution had been spun down and added to 2 μL GlycoBuffer 2 (10× dilution), 2 μL 10% NP‐40 and 6 μL double distilled water to reach a total reaction volume of 20 μL. The mixture was incubated at 37°C for 1 hr and checked by western blotting.
S affinity histochemistry and FCoV immunohistochemistry
Formalin‐fixed, paraffin‐embedded tissues from two mixed breed, male cats diagnosed with mixed type of wet and dry forms of serotype I FIP; and one Chinchilla and two mixed breed 6–10 year‐old cats definitively diagnosed with a disease other than FIP, were sectioned at a thickness of 4 µm. The tissue sections were deparaffinized with xylene and rehydrated with graded ethanol, after which antigens were retrieved by boiling in 5% Trilogy (diluted with distilled water; Cell Marque, Rocklin, CA, USA) for 10 min. Endogenous peroxidase was inactivated by 3% hydrogen peroxide in methanol for 15 min at RT and the slides washed three times with TBST. The slides were blocked with 10% FBS (Gibco) for 30 min at RT and incubated overnight at 4°C with FIPV UU4 S protein or FECV UU7 S protein (5 mg/L, diluted with 1% FBS) or a previously expressed porcine aminopeptidase N (pAPN)‐V5 tagged protein (5 mg/L, diluted with 1% FBS) 32 as a negative control for staining. After FIPV UU4 S protein or control protein affinity binding, the slides were rinsed three times with TBST and incubated with 1000× diluted mouse anti‐V5 Tag antibody (Life Technologies, Carlsbad, CA, USA), in 1% FBS for 1 hr at RT. For FIPV antigen detection, the slides were incubated with a mouse anti‐FCoV antibody (FIPV3‐70; Bio‐Rad) for 1 hr at RT. After washing with TBST, binding was visualized using Dako EnVision+ System‐HRP 3,3′‐diaminobenzidine (Glostrup, Denmark). After washing with TBST, the slides were counterstained with hematoxylin and then mounted with Malinol NX (Muto Pure Chemicals, Tokyo, Japan). To test the role of sialic acid on the interaction of S protein with host cells, sialic acids were removed from the tissues by incubation of the slides with 1 mU NA from Clostridium perfringens (N‐2876, 10 units; Sigma‐Aldrich, St Louis, MO, USA) for 20 hr at 37°C before the S protein histochemistry blocking step. The same S affinity procedure was performed after washing three times with TBST. Binding of the S protein to slides with and without NA treatment was compared using routine light microscopic procedures.
RESULTS
Expression and purification of S proteins
After plasmid transfection of HEK293 cells and cell selection with a high concentration of Geneticin, expressions of the ectodomain of FIPV UU4 and FECV UU7 S proteins were determined by western blotting (Fig. 1b). Under denaturing conditions, the molecular weights of both FIPV UU4 and FECV UU7 S ectodomains were approximately 250 KDa (Fig. 1b). Before deglycosylation, the expressed S proteins bands were smeared because of the polydispersive nature of the glycan chains. After being treated with PNGase F to remove N‐glycans, the bands became sharper, the intensity of signals decreased, and the molecular weights of both FIPV UU4 S and FECV UU7 S ectodomains were reduced to approximately 170 KDa (Fig. 1b), suggesting that the S protein ectodomains produced in the HEK293 cells were highly glycosylated.
Binding of FIPV S protein to paraffin‐embedded tissues
To investigate the binding characteristics of S protein with host cells, S affinity histochemistry was performed by probing recombinant FIPV UU4 or FECV UU7 S protein ectodomains on formalin‐fixed, paraffin‐embedded tissues from cats diagnosed with FIP or cats diagnosed with other diseases. After application of soluble recombinant FIPV UU4 S protein to slides, binding of the protein to large round cells, characterized by a single round to kidney‐bean‐shaped nucleus with abundant cytoplasm resembling tissue macrophages, was observed in various organs (Fig. 2). FIPV UU4 S protein bound macrophages were detected in lesions of cats with FIP and in cats diagnosed with other diseases (Figs. 2 and 3). Less intensive, mild non‐specific binding of FIPV UU4 S protein to cardiac muscle, renal tubular cells, pancreatic acinar cells and hepatocytes was also observed (Fig. 4). As to FECV UU7 S protein affinity histochemistry, absence of FECV UU7 S protein binding on formalin‐fixed, paraffin‐embedded tissues was noted in all tissue slides.
Figure 2.
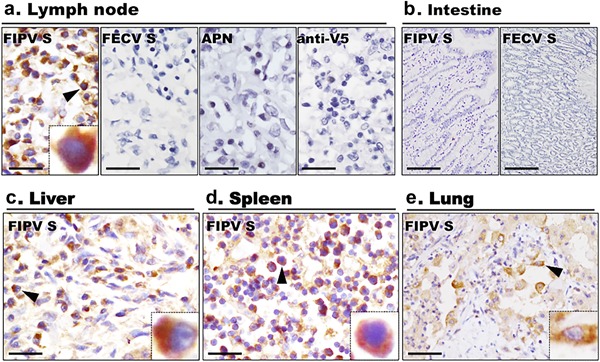
S protein histochemical detection of possible receptors for FCoV I in feline tissues. Using FIPV UU4 S protein (FIPV S) as a probe, binding of S protein to macrophages in (a) lymph node, (c) liver, (d) spleen and (e) lung tissues was noted. No signal was detected in slides incubated with FECV S, recombinant V5‐tagged porcine APN or anti‐V5 tag mAb alone (anti‐V5). (b) No signal was detected on enterocytes in FIPV S or FECV S histochemistry. Scale bars: a, c, d, e = 25 µm; b = 100 µm. Insets showed positive staining under 1,600× magnification.
Figure 4.

S histochemical detection of feline tissues with or without NA treatment. Using FIPV UU4 S protein as a probe, without NA treatment, slight binding of S protein on renal (a) tubular cells, (b) cardiac muscle, (c) pancreas and (d) hepatocytes was detected. With NA treatment, no binding of S protein on (e) renal tubular cells, (f) cardiac muscle, (g) pancreas and (h) hepatocytes was observed. Scale bars: 100 µm.
To investigate the contribution of sialic acid to attachment of serotype I FIPV S protein to feline tissues, slides were pre‐treated with NA to cleave sialic acid from cell surfaces before performing S histochemistry. Interestingly, while binding of FIPV UU4 S protein to macrophages was not affected by pre‐treatment of NA (Fig. 3), binding of S protein to feline hepatocytes, renal tubular cells, cardiac muscle cells and pancreatic cells were completely abolished in the relevant feline tissues pre‐treated with NA (Fig. 4). As to intestinal tissues, regardless of whether or not they were NA treated, there was no evidence of FIPV UU4 or FECV UU7 S protein binding to enterocytes (Fig. 2b). Furthermore, no binding of S protein was observed in slides incubated with the affinity histochemistry negative control pAPN‐V5‐His protein or the antibody control anti‐V5 Tag antibody alone (Fig. 2a).
Figure 3.
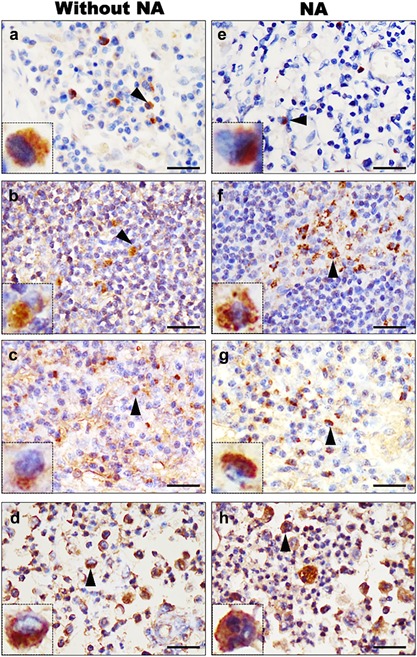
S histochemical detection of possible receptors for FIPV UU4 on cells in lymph nodes of cat diagnosed with FIP (a and e) or cats diagnosed with other disease (b–d, f–h), with or without NA treatment. Using FIPV UU4 S protein as a probe, binding of serotype I FIPV S protein to tissue macrophages was detected. Scale bars: 25 µm. Insets show areas of positive staining of cells indicated by arrowheads under 1,600× magnification.
Correlation of FCoV‐infected cells with S bound cells in paraffin‐embedded tissues
To compare tissue distributions of FIPV UU4 S protein‐bound cells and FIPV antigen‐positive cells, S protein‐bound cells were examined and compared with FCoV antigen‐positive cells on continuous serial sections of paraffin‐embedded tissues from the same anatomic regions. Microscopic examination revealed intracytoplasmic staining of FCoV antigens in macrophages by IHC in pyogranulomatous lesions in livers, spleens, kidneys, lymph nodes and leptomeninges from cats with FIP, whereas no staining was observed in their intestinal tracts. A similar pattern of intracytoplasmic positivity in macrophages associated with FIP pyogranulomatous lesions was also observed by FIPV UU4 S protein affinity histochemistry; however, positive signals in macrophages by S histochemistry were weaker than those obtained by IHC using anti‐FCoV antibody (Fig. 5).
Figure 5.
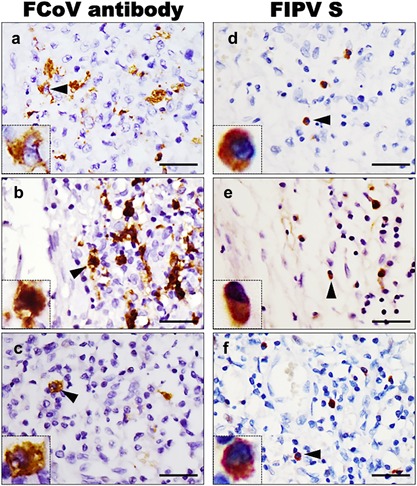
Correlation of FCoV‐infected cells with FIPV S bound cells in serial sections of paraffin‐embedded slides. The distribution of (a, b, and c) FCoV antigens detected by IHC was compared with the distribution of (d, e, and f) FIPV S‐bound cells detected by FIPV UU4 S protein histochemistry. Continuous serial sections of paraffin‐embedded tissues from (a and d) feline liver, (b and e) leptomeninges and (c and f) lymph nodes were incubated with a mouse anti‐FCoV antibody or with FIPV UU4 S protein. Scale bars = 25 µm. Insets show areas of positive signals indicated by arrowheads under 1,600× magnification.
DISCUSSION
In this study, we used a HEK293 expression system to express S protein ectodomains from serotype I FIPV strain UU4 and FECV strain UU7 with the aim of mimicking the attachment of the viral S protein to host cells. We used these highly glycosylated S proteins as a probe for S protein affinity histochemistry and fluorescent staining to investigate the binding characteristics and tissue tropism of serotype 1 FCoVs. The FIPV UU4 S protein primarily bound to macrophages, suggesting a strong macrophage tropism for FIPV strain UU4. Our failure to observe any effect of S protein binding to macrophages after NA pre‐treatment suggests that a factor(s) other than sialic acid is responsible for this tropism of serotype 1 FIPV strain UU4. Furthermore, results of S protein histochemistry and FCoV IHC, which were characterized by positive staining in macrophages, suggest that the tissue distribution of FIPV S protein‐bound cells correlates with that of FIPV antigen‐positive cells and the cell tropism of FIPV.
We established HEK293 cell lines that stably express the ectodomains of S protein derived from FIPV UU4 strain and FECV UU7. However, similar to most glycoproteins, according to western blot examination the molecular weights of these proteins were larger than predicted. After treating S proteins with PNGase F to remove N‐glycans, the molecular weights of both expressed FIPV UU4 and FECV UU7 S ectodomains were reduced to about 170 KDa, suggesting that the S protein ectodomains produced in the HEK293 cells are highly glycosylated. Indeed, using N‐glycosylation site prediction services available from the Center for Biological Sequences 33, at least 13 and 14 N‐glycosylation sites were predicted in the FIPV UU4 S protein and FECV UU7 S protein, respectively. Proteins secreted by mammals often contain obligate post‐translational modifications, including addition of the disulfide bonds and unique glycosylation patterns that are required for proper folding and/or biological activity. We believe that the S protein expressed in the HEK293 system undergoes obligate post‐translational modifications to better bind receptors on cell surfaces. Using the recombinant proteins expressed by the HEK293 system performing S affinity histochemistry on tissues from cats, we demonstrated the binding and interaction of FIPV UU4 S protein with tissue macrophages and feline blood PBMCs. This viral S affinity histochemistry system provides a platform for studying interactions between FIPV binding protein and its host cellular receptors.
To investigate the biological properties of the cellular receptor for FCoV I, we performed S histochemistry with NA pre‐treatment to remove sialic acid from cell surfaces. Sialic acid binding is associated with the pathogenicity of several other coronaviruses. For example, the enteropathogenicity of transmissible gastroenteritis virus in piglets is associated with sialic acid binding of S protein 34, 35, 36. In FCoVs, it has been demonstrated that the sialic acid binding capacity of the S1‐N‐terminal domain can be lost after some serotype I FIPV strains switch to a macrophage tropism 28, and that the S protein of FCoVs appears to be responsible for macrophage tropism 5, 29. In our study, we found that FIPV UU4, but not FECV UU7, was able to bind on the tissue macrophages and that that binding was not affected by NA pre‐treatment. Our results also suggest that a factor(s) other than sialic acid is responsible for the macrophage tropism of serotype I FIPV strain UU4.
As to the application of S affinity histochemistry in intestinal tissues, we failed to identify FIPV UU4 S protein or FECV UU7 S proteins binding on enterocytes. Given that intestinal enzymes cause extremely rapid post‐mortem cellular autolysis of the intestine, the failure of S proteins to bind to enterocytes may be attributable to loss of the membrane integrity of enterocytes. Although the S affinity histochemistry approach used in the present study could serve as a good tool for studying virus–host receptor interactions in paraffin‐embedded tissues, it has limited use in tissues with poor membrane integrity, such as autolytic tissues or intestines. Hence, the present findings concerning interaction of both viral S proteins with their host receptors in the intestine in paraffin‐embedded tissues are inconclusive. Further studies of FECV S affinity histochemistry using feline primary enterocytes or enteric cell lines may reduce the limitations of S affinity histochemistry in paraffin‐embedded tissues.
In the present study, we demonstrated binding of the S protein of serotype I FIPV UU4 to tissue macrophages and showed by staining the same lesions in serial tissue sections that the distribution of S protein‐bound macrophages is related to that of FIPV antigens and lesions associated with FIP. In fact, in our study most tissue macrophages were not recognized by FIPV UU4 S protein. In cats with and without FIP, only some macrophage sublineages bore cellular receptors that were recognized by FIPV UU4 S protein. Indeed, some tissue macrophages are not permissive for FIPV infection, as shown by the failure to detect FIPV antigens in some proliferating macrophage populations, including macrophages in splenic red pulp in cats with FIP infection 37, 38. Future studies aimed at determining which macrophages sublineages are susceptible to FIPV infection would be interesting. Moreover, individual differences in the sustainability and susceptibility of monocytes to infection between cats has been reported 39, indicating that host factors likely also affect interactions between host receptors and S protein binding affinity. We identified fewer FIPV UU4 S than FCoV IHC protein‐bound macrophages/monocytes by S histochemistry. This decreased binding of S protein in affinity histochemistry most likely resulted from tissue damage during slide preparation or loss of cell integrity in pyogranulomatous lesions associated with FIP.
Using S affinity histochemistry, we have here successfully demonstrated the affinity binding of FIPV UU4 S protein to tissue macrophages but not binding of FECV UU7 S protein to these cells. Because tissue damage during slide preparation or loss of cell integrity may have reduced the detection sensitivity of S affinity histochemistry, in the course of our studies we performed S fluorescent staining and flow cytometry to more sensitively and quantitatively detect S protein binding on PBMCs, FCWF‐4 cells and CRFK cells. By using the S fluorescent staining and fluorescent microscopy, we demonstrated that binding of FIPV UU4 S protein to PBMCs occurs in less than 1% of cells (Fig. S1a) and that binding of FECV UU7 S protein on PBMCs was undetectable (Fig. S1b). Using S fluorescent flow cytometry, we detected binding of FIPV UU4 S and FECV UU7 S protein on 15.5% and 7.4% of monocytes gated based on forward scatter and side scatter, respectively (Fig. S1D). We observed no to minimal signals on both FCWF‐4 cells and CRFK cells by S protein affinity fluorescent staining and flow cytometry (Fig. S1C and E). These results indicate that both FIPV UU4 S and FECV UU7 S proteins can recognize small numbers of feline monocytes, which supports that the contention that cell tropism between FIPV and FECV occurs at a post‐receptor step of infection 40. Additionally, S affinity histochemistry may be less sensitive for studying the attachment pattern of FECV I in paraffin‐embedded tissues, which could be attributable to the low cellular receptor/co‐receptor density of FECV I. Moreover, the limited growth of serotype I FCoVs in both FCWF‐4 and CRFK cell lines was possibly caused by the poor binding affinity of FCoV I S protein to these cells.
Using S protein histochemistry on tissue slides, we were able to study and evaluate the interactions of serotype I FIPV binding protein with its host receptor. This assay allowed us to gain more information both on virus–host interactions and the biological characteristics of the unidentified cellular receptor for FIPV I. This approach may be useful for further dissection of S–host interactions, such as identifying the receptor binding domain using variously truncated S proteins, comparing cell tropisms of different pathotypes of FCoVs, identifying virus‐susceptible macrophage sublineages and determining the effects of antibodies or therapeutic compounds on viral S protein binding to target cells.
DISCLOSURE
The authors declare no conflict of interest with respect to the research, authorship and/or publication of this article.
Supporting information
Additional supporting information may be found in the online version of this article at the publisher's web‐site.
Fig. S1. Detection of binding affinity of FIPV UU4 S and FECV UU7 S proteins on feline PBMCs, CRFK cells and FCWF‐4 cells. Freshly prepared feline PBMCs, CRFK cells and FCWF‐4 cells were fixed with 4% paraformaldehyde and resuspended in PBS. The cells were incubated overnight at 4°C with FIPV UU4 S protein, FECV UU7 S protein (5 mg/L, diluted with PBS) or PBS. After incubation, the cells were rinsed with PBS and incubated with anti‐V5 Tag FITC mAb antibody (400× dilution in PBS; Life Technologies) for 1 hr at RT. After washing with PBS, the cells were washed with flow buffer and analyzed by flow cytometry using a FACSCalibur (BD Biosciences) with CellQuest software (BD Biosciences). For cell fluorescent staining, an aliquot of 100 μL of cell suspension was taken from each sample, spun onto glass slides by cytospin centrifuge (Heraeus Instruments, Hanau, Germany), and mounted with VECTASHIELD mounting medium with DAPI (Epitomics, Burlingame, CA, USA). S protein bound cells were imaged with a microscope (Olympus IX‐83; Tokyo, Japan). (a) Detection of binding affinity of FIPV UU4 S on PBMCs, CRFK cells and FCWF‐4 cells by S fluorescent staining. (b) Detection of binding affinity of FECV UU7 S on PBMCs by S fluorescent staining. (c) No signal was detected on PBMCs, Fcwf‐4 cells or CRFK cells using anti‐V5 Tag FITC mAbs alone as the staining control. (d) Histograms show binding of FIPV UU4 S and FECV UU7 S proteins to monocytes. (e) Histograms show binding of FIPV UU4 S and FECV UU7 S proteins to CRFK and FCWF‐4 cells. Binding signals of S protein are in purple. Anti‐V5 Tag FITC mAb antibody was used as a control (bright green).
ACKNOWLEDGMENTS
This work was supported by grant MOST106‐2813‐C‐002‐143‐B from the Ministry of Science and Technology in Taiwan, R.O.C.
REFERENCES
- 1. Fehr A.R., Perlman S. (2015) Coronaviruses: An overview of their replication and pathogenesis. Methods Mol Biol (Clifton, NJ) 1282: 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Calza L., Manfredi R., Verucchi G., Chiodo F. (2003) SARS: A new emergency in the world health. Recenti Prog Med 94: 284–94. [PubMed] [Google Scholar]
- 3. Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. (2012) Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 367: 1814–20. [DOI] [PubMed] [Google Scholar]
- 4. Gorbalenya A.E., Enjuanes L., Ziebuhr J., Snijder E.J. (2006) Nidovirales: Evolving the largest RNA virus genome. Virus Res 117: 17–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Licitra B.N., Millet J.K., Regan A.D., Hamilton B.S., Rinaldi V.D., Duhamel G.E., Whittaker G.R. (2013) Mutation in spike protein cleavage site and pathogenesis of feline coronavirus. Emerg Infect Dis 19: 1066–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R. (2012) Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses 4: 1011–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fiscus S.A., Teramoto Y.A. (1987) Antigenic comparison of feline coronavirus isolates: Evidence for markedly different peplomer glycoproteins. J Virol 61: 2607–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hohdatsu T., Okada S., Koyama H. (1991) Characterization of monoclonal antibodies against feline infectious peritonitis virus type II and antigenic relationship between feline, porcine, and canine coronaviruses. Arch Virol 117: 85–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Horzinek M.C., Lutz H., Pedersen N.C. (1982) Antigenic relationships among homologous structural polypeptides of porcine, feline, and canine coronaviruses. Infect Immun 37: 1148–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Benetka V., Kubber‐Heiss A., Kolodziejek J., Nowotny N., Hofmann‐Parisot M., Mostl K. (2004) Prevalence of feline coronavirus types I and II in cats with histopathologically verified feline infectious peritonitis. Vet Microbiol 99: 31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shiba N., Maeda K., Kato H., Mochizuki M., Iwata H. (2007) Differentiation of feline coronavirus type I and II infections by virus neutralization test. Vet Microbiol 124: 348–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tresnan D.B., Levis R., Holmes K.V. (1996) Feline aminopeptidase N serves as a receptor for feline, canine, porcine, and human coronaviruses in serogroup I. J Virol 70: 8669–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Desmarets L.M., Theuns S., Roukaerts I.D., Acar D.D., Nauwynck H.J. (2014) Role of sialic acids in feline enteric coronavirus infections. J Gen Virol 95: 1911–8. [DOI] [PubMed] [Google Scholar]
- 14. Dye C., Temperton N., Siddell SG. (2007) Type I feline coronavirus spike glycoprotein fails to recognize aminopeptidase N as a functional receptor on feline cell lines. J Gen Virol 88: 1753–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pedersen N.C., Boyle J.F., Floyd K., Fudge A., Barker J. (1981) An enteric coronavirus infection of cats and its relationship to feline infectious peritonitis. Am J Vet Res 42: 368–77. [PubMed] [Google Scholar]
- 16. Pedersen N.C., Black J.W., Boyle J.F., Evermann J.F., McKeirnan A.J., Ott R.L. (1984) Pathogenic differences between various feline coronavirus isolates. Adv Exp Med Biol 173: 365–80. [DOI] [PubMed] [Google Scholar]
- 17. Pedersen N.C. (1976) Morphologic and physical characteristics of feline infectious peritonitis virus and its growth in autochthonous peritoneal cell cultures. Am J Vet Res 37: 567–72. [PubMed] [Google Scholar]
- 18. Petersen N.C., Boyle J.F. (1980) Immunologic phenomena in the effusive form of feline infectious peritonitis. Am J Vet Res 41: 868–76. [PubMed] [Google Scholar]
- 19. Weiss R.C., Scott F.W. (1981) Pathogenesis of feline infetious peritonitis: Pathologic changes and immunofluorescence. Am J Vet Res 42: 2036–48. [PubMed] [Google Scholar]
- 20. Brown M.A., Troyer J.L., Pecon‐Slattery J., Roelke M.E., O'Brien S.J. (2009) Genetics and pathogenesis of feline infectious peritonitis virus. Emerg Infect Dis 15: 1445–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brown M.A. (2011) Genetic determinants of pathogenesis by feline infectious peritonitis virus. Vet Immunol Immunopathol 143: 265–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chang H.W., de Groot R.J., Egberink H.F., Rottier P.J. (2010) Feline infectious peritonitis: insights into feline coronavirus pathobiogenesis and epidemiology based on genetic analysis of the viral 3c gene. J Gen Virol 91: 415–20. [DOI] [PubMed] [Google Scholar]
- 23. Chang H.W., Egberink H.F., Rottier P.J. (2011) Sequence analysis of feline coronaviruses and the circulating virulent/avirulent theory. Emerg Infect Dis 17: 744–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chang H.W., Egberink H.F., Halpin R., Spiro D.J., Rottier P.J. (2012) Spike protein fusion peptide and feline coronavirus virulence. Emerg Infect Dis 18: 1089–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pedersen N.C., Liu H., Dodd K.A., Pesavento P.A. (2009) Significance of coronavirus mutants in feces and diseased tissues of cats suffering from feline infectious peritonitis. Viruses 1: 166–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Poland A.M., Vennema H., Foley J.E., Pedersen N.C. (1996) Two related strains of feline infectious peritonitis virus isolated from immunocompromised cats infected with a feline enteric coronavirus. J Clin Microbiol 34: 3180–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rottier P.J., Nakamura K., Schellen P., Volders H., Haijema B.J. (2005) Acquisition of macrophage tropism during the pathogenesis of feline infectious peritonitis is determined by mutations in the feline coronavirus spike protein. J Virol 79: 14122–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vennema H., Poland A., Foley J., Pedersen N.C. (1998) Feline infectious peritonitis viruses arise by mutation from endemic feline enteric coronaviruses. Virology 243: 150–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Porter E., Tasker S., Day M.J., Harley R., Kipar A., Siddell S.G., Helps C.R. (2014) Amino acid changes in the spike protein of feline coronavirus correlate with systemic spread of virus from the intestine and not with feline infectious peritonitis. Vet Res 45: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ibricevic A., Pekosz A., Walter M.J., Newby C., Battaile J.T., Brown E.G., Holtzman M.J., Brody S.L. (2006) Influenza virus receptor specificity and cell tropism in mouse and human airway epithelial cells. J Virol 80: 7469–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wickramasinghe I.N., de Vries R.P., Grone A., de Haan C.A., Verheije M.H. (2011) Binding of avian coronavirus spike proteins to host factors reflects virus tropism and pathogenicity. J Virol 85: 8903–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yang C., Chang Y.C., Chang C.Y., Tsai P.S., Jeng C.R., Pang VF., Cheng I.C., Chang HW. (2016) Identification of a highly transfectable cell line permissive to porcine epidemic diarrhea virus infection and replication. Taiwan Vet J 42: 187–93. [Google Scholar]
- 33.Center for Biological Sequences. NetNGkyc 1.0 Server [Cited 7 January 2017] Available from URL: http://www.cbs.dtu.dk/services/NetNGlyc
- 34. Krempl C., Schultze B., Laude H., Herrler G. (1997) Point mutations in the S protein connect the sialic acid binding activity with the enteropathogenicity of transmissible gastroenteritis coronavirus. J Virol 71: 3285–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Krempl C., Laude H., Herrler G. (1998) Is the sialic acid binding activity of the S protein involved in the enteropathogenicity of transmissible gastroenteritis virus? Adv Exp Med Biol 440: 557–61. [DOI] [PubMed] [Google Scholar]
- 36. Schultze B., Krempl C., Ballesteros M.L., Shaw L., Schauer R., Enjuanes L., Herrler G. (1996) Transmissible gastroenteritis coronavirus, but not the related porcine respiratory coronavirus, has a sialic acid (N‐glycolylneuraminic acid) binding activity. J Virol 70: 5634–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kipar A., Kohler K., Leukert W., Reinacher M. (2001) A comparison of lymphatic tissues from cats with spontaneous feline infectious peritonitis (FIP), cats with FIP virus infection but no FIP, and cats with no infection. J Comp Pathol 125: 182–91. [DOI] [PubMed] [Google Scholar]
- 38. Kipar A., Meli M.L., Baptiste K.E., Bowker L.J., Lutz H. (2010) Sites of feline coronavirus persistence in healthy cats. J Gen Virol 91: 1698–707. [DOI] [PubMed] [Google Scholar]
- 39. Dewerchin H.L., Cornelissen E., Nauwynck H.J. (2005) Replication of feline coronaviruses in peripheral blood monocytes. Arch Virol 150: 2483–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Regan A.D., Whittaker G.R. (2008) Utilization of DC‐SIGN for entry of feline coronaviruses into host cells. J Virol 82: 11992–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional supporting information may be found in the online version of this article at the publisher's web‐site.
Fig. S1. Detection of binding affinity of FIPV UU4 S and FECV UU7 S proteins on feline PBMCs, CRFK cells and FCWF‐4 cells. Freshly prepared feline PBMCs, CRFK cells and FCWF‐4 cells were fixed with 4% paraformaldehyde and resuspended in PBS. The cells were incubated overnight at 4°C with FIPV UU4 S protein, FECV UU7 S protein (5 mg/L, diluted with PBS) or PBS. After incubation, the cells were rinsed with PBS and incubated with anti‐V5 Tag FITC mAb antibody (400× dilution in PBS; Life Technologies) for 1 hr at RT. After washing with PBS, the cells were washed with flow buffer and analyzed by flow cytometry using a FACSCalibur (BD Biosciences) with CellQuest software (BD Biosciences). For cell fluorescent staining, an aliquot of 100 μL of cell suspension was taken from each sample, spun onto glass slides by cytospin centrifuge (Heraeus Instruments, Hanau, Germany), and mounted with VECTASHIELD mounting medium with DAPI (Epitomics, Burlingame, CA, USA). S protein bound cells were imaged with a microscope (Olympus IX‐83; Tokyo, Japan). (a) Detection of binding affinity of FIPV UU4 S on PBMCs, CRFK cells and FCWF‐4 cells by S fluorescent staining. (b) Detection of binding affinity of FECV UU7 S on PBMCs by S fluorescent staining. (c) No signal was detected on PBMCs, Fcwf‐4 cells or CRFK cells using anti‐V5 Tag FITC mAbs alone as the staining control. (d) Histograms show binding of FIPV UU4 S and FECV UU7 S proteins to monocytes. (e) Histograms show binding of FIPV UU4 S and FECV UU7 S proteins to CRFK and FCWF‐4 cells. Binding signals of S protein are in purple. Anti‐V5 Tag FITC mAb antibody was used as a control (bright green).


