Figure 1. Reconstructing the Architecture of ecDNA using short-read sequencing data.
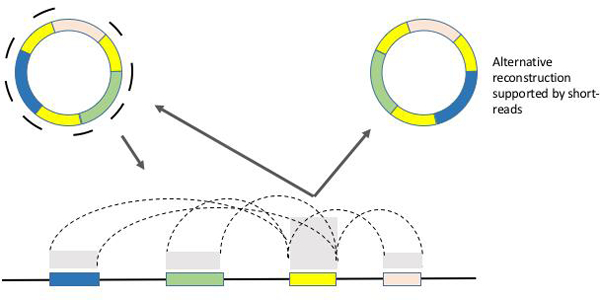
Directed assembly techniques have been developed to reconstruct the architecture of ecDNA. Short-read sequences acquired from ecDNA containing samples are mapped back to a reference genome. The copy number of the reads mapping to ecDNA segments (colored boxes) is proportional to the average number of ecDNA copies per cell, and reveals itself as copy number amplification in the mappings. Reads that span junctions of adjoining segments map back to the reference in a split fashion, revealing breakpoints. These mapping signatures of ecDNA derived complex rearrangements cannot be easily distinguished from chromosomal structural variants, posibly leading to an undercounting of ecDNA. ecDNA reconstruction algorithms explore amplicon graphs where nodes correspond to amplified segments, and edges correspond to breakpoints. Cyclic paths reconstructed by traversing the graph are indicative of ecDNA (Deshpande). Note also that if ecDNA contains large, repeated segments, long reads may be required for unambiguous reconstruction. Improvements in genomic and computational technologies will enhance ecDNA discovery.
