Abstract
Salmonella enterica serovar Choleraesuis is the aetiological agent of swine paratyphoid being a highly invasive zoonotic pathogen. Wild boar natural populations are experiencing a demographical expansion as well as some farms are breeding this species to release for hunting with management sometimes identical to that of domestic pigs, including supplementation, grouping, and antibiotic treatments. This situation increases the chance of contact between wild boars and livestock, and potentially induces stress, with different sanitary consequences. The present work aims to describe the clinical features of recent outbreaks caused by S. Choleraesuis in wild boar from central‐western Spain, as well as the antimicrobial resistance and phylogenetic relationships of isolates involved. 28 strains of S. Choleraesuis were isolated from 28 different wild boars belonging to 10 different game states located in central western Spain and submitted to the Clinical Veterinary Hospital (CVH) of the University of Extremadura. Samples were taken from different organs and cultured according to the ISO 6579:2002 procedure. Suspicious colonies were identified by PCR and antimicrobial resistance was evaluated by disc diffusion susceptibility test and the presence of the main resistance genes as well as 18 plasmid replicons frequently found among the Enterobacteriaceae was verified by PCR. Pulsed field gel electrophoresis was applied to determine the genetic relationship between isolates. The outbreaks under study were characterized by high mortality (35%–84%) and a septicaemic presentation. S. Choleraesuis was isolated from all the wild boars analysed, and 26 of the 28 isolates presented resistance to at least one antibiotic. The predominant resistances found were against sulphonamide, streptomycin, tetracycline, and doxicicline and sul1, strA‐strB, and tetA were the most prevalent resistance genes among isolates. 10 strains carried FIIA, FIB+H/1 or FIIA+H/1 plasmids. PFGE classified the isolates into four different profiles, grouped into two clusters. This results show that prevention against S. Choleraesuis must be considered in the sanitary programs of the wild boar breeders.
Keywords: antibiotic resistance, epidemiology, Salmonella Choleraesuis, wild boar
1. INTRODUCTION
Salmonella Choleraesuis is an intracellular facultative pathogen highly adapted to its host, agent of swine paratyphoid with clinical features of enterocolitis and septicaemia (Reed, Olander, & Thacker, 1986). Although infections in humans are unusual, they can be particularly severe if when occur (Cherubin, 1980). During the 1950s and 1960s, S. Choleraesuis was the predominant serovar in pigs all over the world and although it is still very common in North America and Asia, it is rarely detected in Australia and Western Europe (Fedorka‐Cray, Gray, & Wray, 2000). Most cases reported in Europe came from Estonia and Romania (EFSA, 2015).
However, despite the low prevalence in pigs, S. Choleraesuis is becoming more prevalent in wild boars from Europe, whose population has increased during the last decades (Massei et al., 2015). In some areas of south‐central Spain, the management of the wild boars populations for hunting purposes, including feeding and sometimes estate fencing, increases the risk of occurrence and transmission of diseases (Gortázar, Acevedo, Ruiz‐Fons, & Vicente, 2006). Regarding Salmonellosis, several outbreaks have been reported in Europe in recent years. For example, septicaemic processes, very similar to those described in pigs, were reported in Germany and Italy between the years 2006 and 2013 (Conedera et al., 2014; Methner, Heller, & Bocklisch, 2010).
It has been suggested that a variety of stressors, including the presence of viral disease, could trigger or exacerbate the clinical outbreaks of salmonellosis (Schwartz, 1991). Specifically, most of the studies confirmed the coinfection with the porcine reproductive and respiratory syndrome virus PRRSV (Wills et al., 2000) and the porcine coronavirus type 2 PCV2 (Ha, Jung, Kim, Choi, & Chae, 2005; Lipej et al., 2007; Schulze et al., 2003). However, little is known about how these stressors would affect the course of the disease in wild boars.
From a sanitary and public health perspective, wild boars can play a prominent role in the interplay between Salmonella, livestock and the human population (Hilbert, Smulders, Chopra‐Dewasthaly, & Paulsen, 2012). The continuous growth experienced in game meat consumption and the growing wild boar population density, are helping to increase the chances of disease transmission. Furthermore, it has been shown that 44% of the Salmonella samples isolated from meat or veterinary sources carried resistance to at least one type of antibiotic (Foley & Lynne, 2008). This fact is of major importance, as it is assumed that humans and livestock may be sources for antimicrobial resistance in wildlife (Mentaberre et al., 2013; Navarro‐Gonzalez et al., 2012).
Despite the above stated relevance, little is known about the epidemiology, symptoms, triggering factors (PCV2 & PRRS), or antibiotic resistance profile of Salmonella in wild boars. This work aims to describe the clinical features of recent outbreaks caused by Salmonella Choleraesuis in wild boars from south western Spain, as well as its antimicrobial resistance and phylogenetic relationships.
2. MATERIALS AND METHODS
2.1. Sample collection
Between 2010 and 2016, 28 strains of S. Choleraesuis were isolated from 28 different dead wild boars at the University of Extremadura Clinical Veterinary Hospital (CVH). Biological samples from these animals were sent to the CVH by a hunting management company (Ingulados S.L.). Whole carcasses of 16 of the 28 affected wild boars were received, whereas in the rest of cases only aseptically obtained samples were received, consisting of the main organs of the dead animals (lung, liver, spleen, kidney) and faecal samples, were received. Al animals/samples, were stored at 4°C and sent to CVH within the first 24 hr until analysis.
2.2. Game estates
Wild boars came from 10 different game estates located in the central western zone of the Iberian Peninsula (Figure 1). Most of them were fenced estates (6/10), characterized by a perimeter hunting fence to avoid the pass or scape of animals. Game estate managers generally use cereals or feed as supplements to the natural diet for their wild boars, mostly during the summer. Additionally, two estates were open, with no fences at all, which allow the animals to move freely, where no supplementation is provided. And finally, the remainder 2 (F1 and F10) are considered as “hunting farms”. They are larger estates with effective perimetral and internal fencing. Internal fences isolate an area which serve as breeding centre, supplying wild boars for hunting in specific areas of the same estate or to be sold and transported for hunting in different ones. The managing system in those farms is semi‐intensive, but there are several differences between them. F1 is a 15 Ha farm divided into three identical plots. Plot number one contains 40 females, aged 4. Plot number two contains eight males also aged 4, and the last plot contains the weaned piglets. The animals are vaccinated only against Aujeszky's disease, as this is the only compulsory vaccination for this type of farms. The water is chlorinated and available in drinking troughs, although there were also places where rainwater accumulates. In contrast, F10 farm is divided only into two plots; a small one of approximately 2 Ha and another of 10 Ha. The first plot contained a mixed group of 120 wild boar piglets aging from 2.5 to 6 months that had been previously captured from the bigger plot. After an adaptation period, all the animals ageing 1 year old were moved to the bigger plot. F10 managers did not implement any vaccination protocol and the water supply was not chlorinated in this farm.
Figure 1.
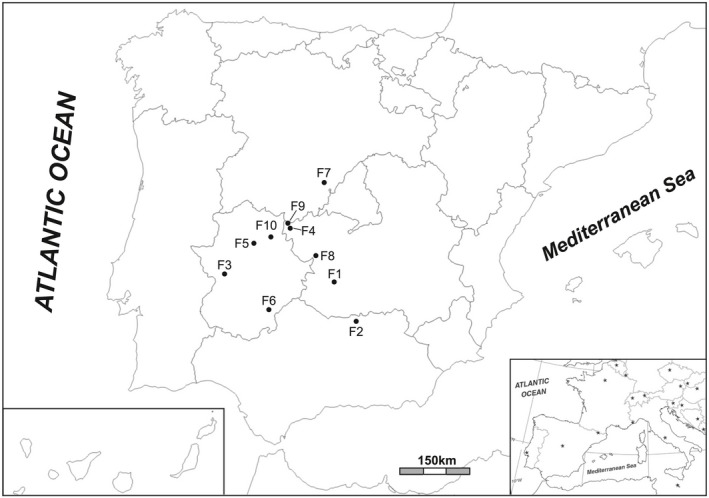
Location of the game estates. Political map of the Iberian Peninsula displaying the location of the different estates investigated in this study. (Inset: Location of the Iberian Peninsula in southwestern Europe)
A summary of the main characteristics of the different estates is shown in Table 1.
Table 1.
Game lands characteristics
| Label | Handling system | Area (ha) | Density (Animals/100 ha) | Livestock presence (Cattle) | Outbreak date (No. animals studied) |
|---|---|---|---|---|---|
| F1 | Game farm with open land * | 15† | 826‡ | No |
June 2010 (7) October 2010 (2) |
| F2 | Open land | 400 | 80 | No | October 2010 (1) |
| F3 | Fenced | 600 | 40 | Yes | May 2011 (1) |
| F4 | Fenced | 2000 | 40 | No |
July 2011 (2) November 2015 (1) |
| F5 | Open land | 1000 | 45 | Yes | June 2012 (1) |
| F6 | Fenced | 700 | 100 | No | July 2014(1) |
| F7 | Fenced | 550 | 90 | No |
July 2013 (1) July 2014 (2) |
| F8 | Fenced | 4500 | 18 | No |
March 2013 (1) November 2015 (1) |
| F9 | Fenced | 3000 | 25 | No |
June 2015 (1) June 2016 (1) |
| F10 | Game farm with open land * | 12† | 1000‡ | No |
July 2015 (2) April 2016 (1) May 2016 (1) June 2016 (1) |
†Fenced section of the estate occupied by the game farm. ‡Animal density based only on the fenced surface occupied by the game farm.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
2.3. Postmortem examination
A complete necropsy was carried out in all the carcasses received. Tissue samples from lungs, liver, spleen, kidneys, and intestine were collected for later histopathological examination. These samples were fixed in 10% neutral buffered formalin and imbibed in paraffin. Tissue sections were cut at 4 μm, stained with H‐E and examined under the microscope.
The age of the animals was determined by the tooth eruption and replacement pattern and also by dental attrition (Boitani & Mattei, 1992).
2.4. Bacteriological culture and identification
All samples taken from the different organs (lungs, liver, kidneys, and spleen) were cultured on blood agar, McConkey agar, and xylose‐lysine‐desoxicholate agar (XLD) in aerobial conditions for 24 hr at 37°C. Faecal samples were pre‐enriched with peptone water (18–24 hr/37°C), enriched in Rappaport‐Vassiliadis Salmonella broth for 48 hr at 42°C, and later cultured in xilose‐lysine‐tergitol 4 (XLT4) and XLD for 48 hr. All procedures were carried out in accordance with ISO 6579:2002/Amd 1:2007 method for the detection of Salmonella.
Identification of compatible colonies was performed suing the Phoenix 100 (Becton Dickinson) automated bacterial identification device and confirmed by detection of the invA gen by PCR (Hoorfar, Ahrens, & Rådström, 2000). PCR‐confirmed isolates were sent to the National Reference Laboratory for Salmonella (Algete, Madrid, Spain) for Kauffman‐White serotyping.
2.5. Antimicrobial resistance, identification of antimicrobial resistance genes, and plasmid typing
The susceptibility testing method used was the antimicrobial disc diffusion susceptibility test in agar recommended by the Clinical and Laboratory Standards Institute (Cockerill, 2011) using 14 antimicrobials from different families which are routinely used in farms. The following discs (Bio‐Rad®) were used: ampicillin (10 μg); cefoxitin (30 μg); ceftiofur (30 μg) gentamicin (10 μg); neomycin (30 μg); streptomycin (10 μg); tetracycline (30 μg); doxycycline (30 μg); enrofloxacin (5 μg); nalidixic Acid (30 μg); trimethoprim/sulphamethoxazole (23,75/1,25 μg); sulphonamide (200 μg); chloramphenicol (30 μg); colistin (50 μg). Escherichia coli ATCC 25922 was used as control strain.
The presence of antimicrobial resistance genes was verified by specific PCRs for genes bla‐TEM, bla‐OXA, tet(A), tet(B), aadA, strA, strB, and sul1 (Aarestrup et al., 2003). Isolates were also examined for the presence of the 18 plasmid replicons frequently found among the Enterobacteriaceae, using three multiplex panels (Johnson et al., 2007). Positive controls used in the replicon typing procedure were kindly provided by Alessandra Carattoli (Istituto Superiore di Sanità, Rome, Italy).
2.6. Phylogenetic analysis using pulsed‐field gel electrophoresis (PFGE)
Determination of the genetic relationship between isolates was performed by macrorestriction with XbaI followed by PFGE (Chef‐DR®III. BioRad®), according to the PulseNet protocol with pulse oscillated from 2.16 to 63.8 s for 21.5 hr (Ribot et al., 2006). The different PFGE profiles (PFPs) were analysed by InfoQuest FP Software (Version 4.5).
2.7. Porcine circovirus type 2 (PCV‐2) and Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) analysis
Only 14 blood samples could be collected directly from heart cavities, preserved in refrigeration until centrifugation at 1500 g for 5 min. Serum obtained from each sample was stored at −20°C until use. To determine the presence of antibodies against PCV‐2 and PRRSV, samples were analysed by commercial kits of enzymatic immunoassay technique (ELISA) (Ingezim Circovirus IgG/IgM and Ingezim PRRS Universal), following the manufacturer instructions (Ingenasa, Madrid, Spain).
3. RESULTS
3.1. Sample origin: outbreaks description
Farms F1 and F10 have a semi‐intensive management system, which allows a close monitoring of the animals and almost daily checking of all individuals. The rest of the samples were collected in estates without that exhaustive control of the livestock, making impossible to obtain accurate epidemiological data from those places. The age of the animals from those estates was estimated at around 2–5 months.
The outbreaks occurred in the estates F1 and F10. In F1, symptoms started 1 week after weaning, during June 2010, when a few wild boar piglets around 3 months‐old from the common growing fence (Figure 2a) displayed anorexia and depression. Two days later, 15 wild piglets suddenly died with no symptoms and the rest were getting subsequently sick. The affected animals showed anorexia, depression, aqueous‐greenish diarrhoea, walking difficulties, and finally prostration. The course of the infection in these animals lasted 2–3 days and, at the end, most of the diseased animals died, thus reaching a 100% morbidity with an 84.5% (68/76) mortality. The duration of this outbreak was approximately 2–3 weeks.
Figure 2.
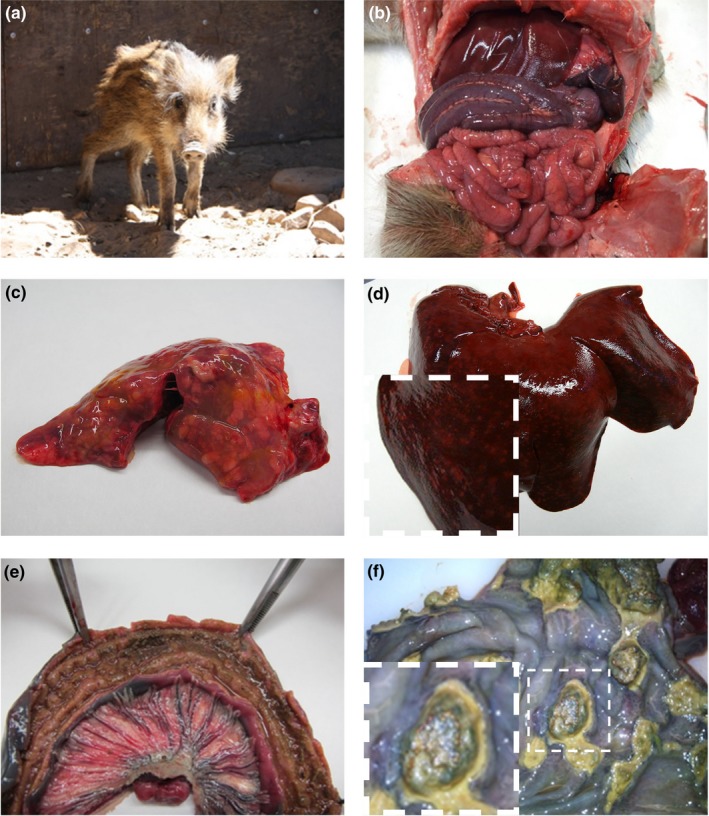
Clinical symptoms and pathological findings. (a) Wild boar piglet with poor corporal condition. (b) Intense congestion observed in the abdominal cavity from a wild boar piglet. (c) Congestive lung displaying multiple pneumonic lesions. (d) Liver showing hepatomegaly and congestion. Inset: Magnification of the border of the right medial hepatic lobe displaying white spots in the parenchima. (e) Longitudinally opened jejunum section revealing a thickened mucosa and a dark and gritty content. (f) Colonic mucosa with multiple ulcers. Inset: Magnification of one of the colonic ulcers [Colour figure can be viewed at http://wileyonlinelibrary.com]
The outbreak in F10 occurred in July 2015, after the capture of 120 wild boar piglets ranging from 2.5 to 6 months of age. In this case, 32 animals died in the first 2 days (not showing previous clinical symptoms) and 10 more during the following week (displaying a profuse diarrhoea). The morbidity reached the 80% of the herd and the mortality was 35% (42/120).
3.2. Pathological findings in wild boars
Externally, most carcasses showed distal cyanosis, especially in ears, legs, and lower part of the abdomen. All organs exhibited diffuse congestion (Figure 2b). The lungs presented pneumonic lesions affecting either the cranial or the apical lobes (Figure 2c). In the abdomen, the most common findings were hepatomegaly (Figure 2d) and splenomegaly, frequently accompanied by small white spots (≈2 mm) in the hepatic parenchyma. Renal petechial spots were also observed in some animals. Regarding the intestine, the most frequent lesion was a mesenteric lymphadenitis as well as a congestion of the mesenteric vessels (Figure 2e). Enteritis and colitis were found in some animals. The jejune and ileum from those animals showed a thickened mucosa covered by a brown fibrinous membrane (Figure 2e) and two of them presented well‐defined rounded ulcers in colon and caecum (Figure 2f). The intestinal content was dark and gritty in most of the specimens.
Microscopically, an interstitial pneumonia was observed in the lung, together with oedema and congestion. The liver displayed interstitial nonpurulent hepatitis, with areas of cellular necrosis all over the hepatic parenchyma. In the spleen an increase in the white pulp was observed and the kidneys showed congestion with tubulonephrosis and interstitial nephritis.
3.3. Isolation and characterization of S. Choleraesuis
A septicaemic process was detected in all animals under study. The bacterial analysis performed to the samples showed that only 4 of 28 animals (14.2%) were also excreting salmonella when they died, as the samples taken from the intestine of those animals resulted positive.
Every one of the 28 bacterial strains, isolated from different animals, were classified as S. enterica subsp. enterica and their biochemical profiles and antigenic formulae were consistent with S. Choleraesuis, although more than a half (16/28) lacked the first flagellar antigen. The Kunzendorf variant was detected in 22 isolates, six presenting both flagellar antigens (Figure 3).
Figure 3.

Phylogenetic relationship among 28 isolates of Salmonella enterica serotype Choleraesuis obtained from wild boars from the south‐central part of the Iberian Peninsula, listed with additional information about the date and place of the isolate as well as its genotypic and phenotypic resistance profile, plasmid presence and serological analysis to the presence of PCV‐2 and PRRSV. Dendogram shows four different profiles (SC1‐SC4) further clustered in groups A and B. Phenotipical resistance pattern: Su (sulphonamide); S (streptomycin); Te (tetracycline); Do (doxicycline); Na (nalidixic acid); N (neomycin); Sxt (trimethoprim/sulfamethoxazole); Am (ampicillin). Serological analisys: Neg (Negative); ‐ (Not tested). (Blanks represent absence of resistance or plasmids)
According to clinical breakpoints, 26 of the 28 strains were resistant to at least one antibiotic. Amongst these, 11 isolates showed resistance to 3–5 antibiotics and could be considered multiresistant. Regarding antimicrobial groups, predominant resistances are found against sulphonamide (n = 22), streptomycin (n = 18), tetracycline (n = 12), and doxicycline (n = 10). Finally, there were also two resistant strains against nalidixic acid and three with a single resistance, to ampicillin, neomicyn, and trimethoprim‐sulfamethoxazole, respectively (Figure 3). All isolates were susceptible to ceftiofur, cefoxitin, gentamicin, enrofloxacin, chloramphenicol, and colistin. sul1, strA‐strB, and tetA genes encoding antimicrobial resistance against sulphonamide, streptomycin, and tetracycline, respectively, were highly prevalent amongst isolates and closely linked to their corresponding resistant phenotypes, being SuSTeDo and SuS the major patterns with 9 and 6 isolates, respectively. Other antimicrobial resistance determinants were marginally observed, like tetB or aadA genes, each one in one different isolate and bla‐TEM in the unique ampicillin resistant isolate amongst all the screened S. Choleraesuis. In addition, replicon typing detected 10 strains carrying plasmids, namely FIIA (n = 1), FIB+H/1 (1), or FIIA+H/1 (8) (Figure 3).
PFGE identified four different profiles, SC1‐SC4, which are further clustered in groups A and B (Figure 3). SC1 and SC2 are represented by 10 strains coming from three different estates closely located: F4, F9, and F10 (Figures 1 and 3). Even higher is the similarity detected among SC3 and SC4 isolates from seven different estates lacking geographical connection.
3.4. Serological analyses
The fourteen sera analysed were negative to PRRSV, although two of them resulted positive to PCV‐2. These two samples belonged to the same farm (F10) and outbreak (July, 2015), and showed high levels of IgM and IgG, indicating an acute status of the infection.
4. DISCUSSION
In this work, we described the occurrence of multiple cases of septicaemic salmonellosis in young wild boars from game estates located in central‐western Spain. The outbreaks reported in this study were characterized by remarkably high mortality rates, 35%–84.5%, much more elevated than the 10% reported in the only case known to date for farmed wild boars in Spain (Pérez et al., 1999), and more similar, but still higher than the 4.2%–33% rates described in pigs from Denmark and Japan (Murakami et al., 2006; Pedersen et al., 2015). Our results seem to contradict the idea that wild boars act only as reservoirs for Salmonella, being carriers and intermittent shedders (Ruiz‐Fons, 2017). The age of the affected wild boars resulted similar than those described in the literature for pigs or wild boars, being related to the weaning period or other stressing conditions as well (Carlson, Barnhill, & Griffith, 2012; Perez et al., 1999). It should be noted that the highest rates came exclusively from the estates with semi‐intensive management of the animals, F1 and F10, where stressful conditions may have a major impact on the health status of the animals (Giles, Belkhiri, Barrow, & Foster, 2017). This fact, together with the poor specific immunity of wild boars against S. Choleraesuis (Methner et al., 2010), could decrease their natural resistance and even activate dormant S. Choleraesuis (Chiu, Su, & Chu, 2004). In our cases, weaning (F1) and grouping animals of different ages and origins (F10) were probably the situations that triggered the outbreaks. Both measures are likely to induce stress and thus facilitate the exchange and dissemination of pathogens (Giles et al., 2017; Roth & Thacker, 2006). The higher mortality observed in F1 could be due to the fact that at ages of 2‐3 months the piglets show the lowest level of antibodies since birth, being much more susceptible to infections (Tizard, 2009).
The management strategies applied in the hunting areas are critical for the sanitary status of the animals. High animal densities would have a major impact on the hygienic habits of the animals and also on their social stress (Fernández‐Llario, Carranza, & Hidalgo de Trucios, 1996; Morrow‐Tesch, McGlone, & Salak‐Johnson, 1994), facilitating the contagion and development of pathological processes. In addition, it should be considered that initially harmless pathogens such as S. serovar Saintpaul, could produce symptoms of marked virulence in wild boars (Ecco, Guedes, Tury, Santos, & Perecmanis, 2006). In the same way, some common porcine pathogens cause especially high mortalities in this species, e.g., Erysipelothrix rhusiopathiae, Pasteurella multocida, or Staphylocccus hyicus (Risco, Fernandez‐Llario, Cuesta, et al., 2013; Risco, Fernandez‐Llario, Velarde, et al., 2013; Risco et al., 2011). So, while the infective doses of S. Choleraesuis necessary for the onset of the disease in young pigs in natural conditions are >108 UFC/g. (Gray, Fedorka‐Cray, Stabel, & Kramer, 1996), in wild boar, due to their poor specific immunity, these doses could be much smaller (Methner et al., 2010). Altogether, these findings highlight how critical an adequate management of wild boars could be in semi‐intensive conditions.
Many previous reports pointed out the possibility of a triggering effect caused by immunosuppressant viruses (especially PCV‐2) in activating Salmonella disease in pigs (Chiu et al., 2004; Ha et al., 2005; Schwartz, 1991; Wills et al., 2000). Nevertheless, in our samples, only two animals showed high values of both IgG and IgM against PCV‐2, indicating an active status of this viral infection at the time of death. These two animals belonged to the same farm (F10) and outbreak (July, 2015). The rest of the animals resulted negative to the active presence of PCV‐2 or PRRSV. Similar results were also described more recently in Italy (Conedera et al., 2014), where none of the wild boars analysed was positive to those viruses. Taken together, these data suggest that, in wild boar, S. Choleraesuis does not need a previous immunosuppressant infection in order to develop a pathological process.
The clinical symptoms and lesions observed in our animals were comparable to those described in similar processes affecting domestic pigs (Fedorka‐Cray et al., 2000) and wild boars (Conedera et al., 2014; Pérez et al., 1999), with the exception of the distal cyanosis, affecting ears, extremities, and ventral area of the abdomen that had not be previously described. In accordance with the aforementioned reports, the lesions found in our study indicated a septicaemic presentation, as confirmed by the isolation of S. Choleraesuis from nongastrointestinal organs, most notably lungs. This presentation is often related to an inhalatory transmission (Gray, Fedorka‐Cray, Stabel, & Ackermann, 1995) which have been shown to be more frequent than oral (Clemmer, Hickey, Bridges, Schliessmann, & Shaffer, 1960), especially in intensive breeding farms and in the summer months, due to the dryness of soil (Baskerville & Dow, 1973). In our study, 18 of the 28 samples were collected in summer, similar to all the outbreaks reported previously in this species (Methner et al., 2010; Perez et al., 1999), suggesting that dust and aerosols generated by sneezing could have a mayor impact in the transmission and dissemination of the disease in wild boars groups (Fedorka‐Cray et al., 2000).
The S. Choleraesuis isolates found in the animals under study displayed a variety of resistances against different antimicrobials, being sulphonamides and tetracyclines the groups with the greatest percentages of resistant strains. This could be related to the regular use of single sulphonamides or combinations of sulphonamides with tetracyclines in the prophylaxis and treatment of diverse pathologies that historically affected some of the farms in this study. A similar result was also obtained in Danish pig herds, in the only study reported to date in Europe about antibiotic resistance in S. Choleraesuis from pigs (Pedersen et al., 2015). Studies from United States showed higher rates of resistance to tetracyclines (92.6%) (Huang, Lin, & Wu, 2009) while data from Japan revealed multidrug resistant isolates with resistance to fluoroquinolones and cephalosporines (Asai et al., 2010; Chang et al., 2005; Chiu et al., 2004). Other screening performed in wild boars from Germany revealed high resistance of S. Choleraesuis to sulphamethoxazole and streptomycin (Methner et al., 2010) whereas very high rates of resistance against streptomycin (73%), spiramicin, and tilmicosin (both at 100%) have been found in Italy (Donazzolo et al., 2017). To date, the present study is the only report that analyses the resistance genes and the plasmids replicons present in S. Choleraesuis isolates from wild boars.
The estates belonging to cluster A are located in same geographical region, specially F4 and F9, which were adjacent lands with no physical barriers between them and similar feeding management, commonly supplemented with sulphonamides. This fact explains the high degree of similarity in PFGE (SC2) or phenotypical resistance patterns (SuSTeDo) found in the strains from these places. The other estate from cluster A, F10, was located 44 km away from F4 and F9 but still in the same region. As mentioned above, F10 is a “hunting farm” that provides animals to other places and implements sanitary measures including antibiotics administration. F10 shared identical phenotypical resistance pattern with F4 and F9 but displayed the PFGE profile SC1. It should be mentioned that SC1 profile also appeared in F4 a few years later after the first outbreak was declared in this estate (Isolate R144), clearly supporting the hypothesis of genetic transfer between estates of this cluster. Regarding to cluster B, most of its isolates were classified into SC4. All the strains from this subgroup (SC4) lacked the flagellar antigen and presented the Kunzendorf variant. Besides, its phenotypical resistance pattern was much more limited than that from cluster A. Unlike SC4, strains from SC3 presented the flagellar antigen and showed different resistant patterns (genotypical and phenotypical) which could be due to a different evolution in their ecological niches, acquiring distinct resistances. Despite the high degree of similarity in cluster B (97%), there was no apparent geographical relation between the locations of the isolates in this group. This remarkable dispersion could be due to the ability of the strains of Salmonella to persist for long periods of time in asymptomatic carriers, as it was previously demonstrated in Germany (Methner et al., 2010). Such persistence would explain the distance between isolates as well as the time between different cases.
As concluding remarks, the special virulence observed in our data highlights the importance of the management‐related stressors in this species. The dissemination of S. Choleraesuis triggered by the manipulation of the herd, together with the special susceptibility of this wild animals to the immunosuppressant effect caused by the stress, emphasise the necessity of specific management methods in this species. On the other hand, our results points to a possible relation between human intervention and the presence of higher rates of antibiotic resistance, as it has been previously reported in different wild mammals (Allen et al., 2010). In order to avoid future difficulties with the productive management of wild boars as well as to reduce the human impact on their environment, it is necessary to re‐evaluate the management methods applied on this species. More studies are needed to implement procedures specifically designed for breeding of these wild animals with a minimal interaction from humans.
CONFLICT OF INTEREST
All the authors have read the manuscript and have approved this submission. All have made substantive contributions to this work. The authors report no conflicts of interest.
ACKNOWLEDGEMENTS
Dr. A. Garcia was supported by contract TA13003 granted by Junta de Extremadura and the European Social Fund. Dr. D. Risco was supported by a Torres Quevedo Grant of the Ministerio de Economía y Competitividad of Spain (PTQ14‐06663)
Gil Molino M, Risco Pérez D, Gonçalves Blanco P, et al. Outbreaks of antimicrobial resistant Salmonella Choleraesuis in wild boars piglets from central‐western Spain. Transbound Emerg Dis. 2019;66:225–233. 10.1111/tbed.13003
REFERENCES
- Aarestrup, F. M. , Lertworapreecha, M. , Evans, M. C. , Bangtrakulnonth, A. , Chalermchaikit, T. , Hendriksen, R. S. , & Wegener, H. C. (2003). Antimicrobial susceptibility and occurrence of resistance genes among Salmonella enterica serovar Weltevreden from different countries. The Journal of antimicrobial chemotherapy, 52(4), 715–718. [DOI] [PubMed] [Google Scholar]
- Allen, H. K. , Donato, J. , Wang, H. H. , Cloud‐Hansen, K. A. , Davies, J. , & Handelsman, J. (2010). Call of the wild: antibiotic resistance genes in natural environments. Nature Reviews Microbiology, 8(4), 251–259. [DOI] [PubMed] [Google Scholar]
- Asai, T. , Namimatsu, T. , Osumi, T. , Kojima, A. , Harada, K. , Aoki, H. , & Takahashi, T. (2010). Molecular typing and antimicrobial resistance of Salmonella enterica subspecies enterica serovar Choleraesuis isolates from diseased pigs in Japan. Comparative Immunology, Microbiology and Infectious Diseases, 33(2), 109–119. doi: 10.1016/j.cimid.2008.08.004 [DOI] [PubMed] [Google Scholar]
- Baskerville, A. , & Dow, C. (1973). Pathology of experimental pneumonia in pigs produced by Salmonella cholerae‐suis. Journal of Comparative Pathology, 83(2), 207–215. [DOI] [PubMed] [Google Scholar]
- Boitani, L. , & Mattei, L. (1992). Aging wild boar (Sus scrofa) by tooth eruption In Spitz F., Janeau G., Gonzalez G., & Aulagnier S. (Eds.), Ongulés/Ungulates. Société Française pour l’Étude et la Protection des Mammifères: Paris‐Toulouse. [Google Scholar]
- Carlson, A. , Barnhill, A. E. , & Griffith, R. W. (2012). Salmonellosis In Zimmerman J. J. (Ed.), Diseases of Swine, Vol. 1 (pp. 821–833). West Sussex, UK: John Wiley and Sons Ltd. [Google Scholar]
- Chang, C.‐C. , Lin, Y.‐H. , Chang, C.‐F. , Yeh, K.‐S. , Chiu, C.‐H. , Chu, C. , & Chiou, C.‐S. (2005). Epidemiologic relationship between fluoroquinolone‐resistant Salmonella enterica serovar Choleraesuis strains isolated from humans and pigs in Taiwan (1997 to 2002). Journal of Clinical Microbiology, 43(6), 2798–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherubin, C. (1980). Epidemiologic assessment of antibiotic resistance in salmonella In Steele D. J. (Ed.), CRC Handbook Series in Zoonosis (Vol. I Sect, pp. 173–200). Boca Raton, FL: CRC Press. [Google Scholar]
- Chiu, C. H. , Su, L. H. , & Chu, C. (2004). Salmonella enterica serotype choleraesuis: Epidemiology, pathogenesis, clinical disease, and treatment. Clinical Microbiology Reviews, 17(2), 311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemmer, D. I. , Hickey, J. L. , Bridges, J. F. , Schliessmann, D. J. , & Shaffer, M. F. (1960). Bacteriologic studies of experimental air‐borne salmonellosis in chicks. Journal of Infectious Diseases, 106, 197–210. [DOI] [PubMed] [Google Scholar]
- Cockerill, F. R. (2011). Performance standards for antimicrobial susceptibility testing: twenty‐first informational supplement. Wayne, PA: Clinical and Laboratory Standards Institute (CLSI). [Google Scholar]
- Conedera, G. , Ustulin, M. , Barco, L. , Bregoli, M. , Re, E. , Vio, D. (2014). Outbreak of atypical Salmonella Choleraesuis in wild boar in North Eastern Italy In Paulsen P., Bauer A., & Smulders F. J. M. (Eds.), Trens in game meat hygiene (pp. 151–159). Wageningen: Academic Publishers. [Google Scholar]
- Donazzolo, C. , Turchetto, S. , Ustulin, M. , Citterio, C. , Conedera, G. , Vio, D. , … Cocchi, M. (2017). Antimicrobial susceptibility of Salmonella enterica subsp. enterica serovar Choleraesuis strains forum wild boar (Sus scrofa) in Italy In Bauer A., Paulsen P., F.J.M. & Smulders (Ed.), Game meat hygiene (pp. 307). The Netherlands: Wageningen Academic Publishers. [Google Scholar]
- Ecco, R. , Guedes, R. M. C. , Tury, E. , Santos, H. L. Jr , & Perecmanis, S. (2006). Outbreak of enterocolitic salmonellosis on a wild pig farm. Veterinary Record, 158, 242–243. [DOI] [PubMed] [Google Scholar]
- EFSA (2015). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food‐borne outbreaks in 2014. EFSA Journal, 13, 4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorka‐Cray, P. J. , Gray, J. T. , & Wray, C. (2000). Salmonella infections in pigs In Wray C., & Wray A. (Eds.), Salmonella in domestic animals (pp. 191–207). London: CABI. [Google Scholar]
- Fernández‐Llario, P. , Carranza, J. , & Hidalgo de Trucios, S. (1996). Social organization of the wild boar (Sus scrofa) in Doñana National Park. Miscellania Zoologica, 19, 8–18. [Google Scholar]
- Foley, S. L. , & Lynne, A. M. (2008). Food animal‐associated Salmonella challenges: Pathogenicity and antimicrobial resistance. Journal of Animal Science, 86(14 Suppl), E173–E187. 10.2527/jas.2007-0447 [DOI] [PubMed] [Google Scholar]
- Giles, T. A. , Belkhiri, A. , Barrow, P. A. , & Foster, N. (2017). Molecular approaches to the diagnosis and monitoring of production diseases in pigs. Research in Veterinary Science, 114, 266–272. 10.1016/j.rvsc.2017.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gortázar, C. , Acevedo, P. , Ruiz‐Fons, F. , & Vicente, J. (2006). Disease risks and overabundance of game species. European Journal of Wildlife Research, 52(2), 81–87. 10.1007/s10344-005-0022-2 [DOI] [Google Scholar]
- Gray, J. T. , Fedorka‐Cray, P. J. , Stabel, T. J. , & Ackermann, M. R. (1995). Influence of inoculation route on the carrier state of Salmonella choleraesuis in swine. Veterinary Microbiology, 47(1–2), 43–59. doi: 10.1016/0378-1135(95)00060-N [DOI] [PubMed] [Google Scholar]
- Gray, J. T. , Fedorka‐Cray, P. J. , Stabel, T. J. , & Kramer, T. T. (1996). Natural transmission of Salmonella choleraesuis in swine. Applied and Environmental Microbiology, 62(1), 141–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha, Y. , Jung, K. , Kim, J. , Choi, C. , & Chae, C. (2005). Outbreak of salmonellosis in pigs with postweaning multisystemic wasting syndrome. Veterinary Record‐English Edition, 156(18), 583–584. [DOI] [PubMed] [Google Scholar]
- Hilbert, F. , Smulders, F. J. M. , Chopra‐Dewasthaly, R. , & Paulsen, P. (2012). Salmonella in the wildlife‐human interface. Food Research International, 45(2), 603–608. 10.1016/j.foodres.2011.08.015 [DOI] [Google Scholar]
- Hoorfar, J. , Ahrens, P. , & Rådström, P. (2000). Automated 5′ nuclease PCR assay for identification of Salmonella enterica . Journal of Clinical Microbiology, 38(9), 3429–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, T. M. , Lin, T. , & Wu, C. (2009). Serovar distribution and antimicrobial susceptibility of swine Salmonella isolates from clinically ill pigs in diagnostic submissions from Indiana in the United States. Letters in applied microbiology, 48(3), 331–336. [DOI] [PubMed] [Google Scholar]
- Johnson, T. J. , Wannemuehler, Y. M. , Johnson, S. J. , Logue, C. M. , White, D. G. , Doetkott, C. , & Nolan, L. K. (2007). Plasmid replicon typing of commensal and pathogenic Escherichia coli isolates. Applied and Environmental Microbiology, 73(6), 1976–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipej, Z. , Segales, J. , Jemeršić, L. , Olvera, A. , Roić, B. , Novosel, D. , & Manojlović, L. (2007). First description of postweaning multisystemic wasting syndrome (PMWS) in wild boar (Sus scrofa) in Croatia and phylogenetic analysis of partial PCV2 sequences. Acta Veterinaria Hungarica, 55(3), 389–404. [DOI] [PubMed] [Google Scholar]
- Massei, G. , Kindberg, J. , Licoppe, A. , Gacic, D. , Sprem, N. , Kamler, J. , … Nahlik, A. (2015). Wild boar populations up, numbers of hunters down? A review of trends and implications for Europe. Pest Management Science, 71(4), 492–500. 10.1002/ps.3965 [DOI] [PubMed] [Google Scholar]
- Mentaberre, G. , Porrero, M. C. , Navarro‐Gonzalez, N. , Serrano, E. , Dominguez, L. , & Lavin, S. (2013). Cattle drive Salmonella infection in the wildlife‐livestock interface. Zoonoses Public Health, 60(7), 510–518. 10.1111/zph.12028 [DOI] [PubMed] [Google Scholar]
- Methner, U. , Heller, M. , & Bocklisch, H. (2010). Salmonella enterica subspecies enterica serovar Choleraesuis in a wild boar population in Germany. European Journal of Wildlife Research, 56, 493–502. [Google Scholar]
- Morrow‐Tesch, J. L. , McGlone, J. J. , & Salak‐Johnson, J. L. (1994). Heat and social stress effects on pig immune measures. Journal of Animal Science, 72(10), 2599–2609. [DOI] [PubMed] [Google Scholar]
- Murakami, S. , Ogawa, A. , Kinoshita, T. , Matsumoto, A. , Ito, N. , & Nakane, T. (2006). Occurrence of swine salmonellosis in postweaning multisystemic wasting syndrome (PMWS) affected pigs concurrently infected with porcine reproduction and respiratory syndrome virus (PRRSV). Journal of Veterinary Medical Science, 68(4), 387–391. [DOI] [PubMed] [Google Scholar]
- Navarro‐Gonzalez, N. , Mentaberre, G. , Porrero, C. M. , Serrano, E. , Mateos, A. , Lopez‐Martin, J. M. , & Dominguez, L. (2012). Effect of cattle on Salmonella carriage, diversity and antimicrobial resistance in free‐ranging wild boar (Sus scrofa) in northeastern Spain. PLoS ONE, 7(12), e51614 10.1371/journal.pone.0051614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen, K. , Sorensen, G. , Lofstrom, C. , Leekitcharoenphon, P. , Nielsen, B. , Wingstrand, A. , & Baggesen, D. L. (2015). Reappearance of Salmonella serovar Choleraesuis var. Kunzendorf in Danish pig herds. Veterinary Microbiology, 176(3–4), 282–291. 10.1016/j.vetmic.2015.01.004 [DOI] [PubMed] [Google Scholar]
- Perez, J. , Astorga, R. , Carrasco, L. , Mendez, A. , Perea, A. , & Sierra, M. (1999). Outbreak of salmonellosis in farmed European wild boars (Sus scrofa ferus). Veterinary Record, 145(16), 464–465. [DOI] [PubMed] [Google Scholar]
- Pérez, J. , Astorga, R. , Carrasco, L. , Méndez, A. , Perea, A. , & Sierra, M. A. (1999). Outbreak of salmonellosis in farmed European wild boars (Sus scrofa ferus). Veterinary Record, 145, 464–465. [DOI] [PubMed] [Google Scholar]
- Reed, W. M. , Olander, H. J. , & Thacker, H. L. (1986). Studies on the pathogenesis of Salmonella typhimurium and Salmonella choleraesuis var kunzendorf infection in weanling pigs. American Journal of Veterinary Research, 47, 75–83. [PubMed] [Google Scholar]
- Ribot, E. M. , Fair, M. , Gautom, R. , Cameron, D. , Hunter, S. , Swaminathan, B. , & Barrett, T. J. (2006). Standardization of pulsed‐field gel electrophoresis protocols for the subtyping of Escherichia coli O157: H7, Salmonella, and Shigella for PulseNet. Foodbourne Pathogens & Disease, 3(1), 59–67. [DOI] [PubMed] [Google Scholar]
- Risco, D. , Fernandez‐Llario, P. , Cuesta, J. M. , Garcia‐Jimenez, W. L. , Gil, M. , Goncalves, P. , & Hermoso de Mendoza, J. H. (2013). Fatal outbreak of systemic pasteurellosis in a wild boar (Sus scrofa) population from southwest Spain. Journal of Veterinary Diagnostic Investigation, 25(6), 791–794. 10.1177/1040638713504411 [DOI] [PubMed] [Google Scholar]
- Risco, D. , Fernandez‐Llario, P. , Velarde, R. , Cuesta, J. M. , Garcia, W. L. , Goncalves, P. , & Hermoso de Mendoza, J. H. (2013). A case of exudative epidermitis in a young wild boar from a Spanish game estate. Journal of Swine Health and Production, 21(6), 4. [Google Scholar]
- Risco, D. , Fernandez‐Llario, P. , Velarde, R. , Garcia, W. L. , Benitez, J. M. , Garcia, A. , & Gomez, L. (2011). Outbreak of swine erysipelas in a semi‐intensive wild boar farm in Spain. Transboundary and Emerging Diseases, 58(5), 445–450. 10.1111/j.1865-1682.2011.01234.x [DOI] [PubMed] [Google Scholar]
- Roth, J. A. , & Thacker, E. L. (2006). Immune System In Straw B. E. (Ed.), Diseases of swine (9, th ed. (pp. 15–36). Ames, Iowa: Blackwell Publishing. [Google Scholar]
- Ruiz‐Fons, F. (2017). A review of the current status of relevant zoonotic pathogens in wild swine (Sus scrofa) populations: Changes modulating the risk of transmission to humans. Transboundary and Emerging Diseases, 64(1), 68–88. 10.1111/tbed.12369 [DOI] [PubMed] [Google Scholar]
- Schulze, C. , Neumann, G. , Grütze, I. , Engelhardt, A. , Mirle, C. , Ehlert, F. , & Hlinak, A. (2003). Case report: Porcine circovirus type 2 infection in an European wild boar (Sus scrofa) in the state of Brandenburg, Germany. DTW. Deutsche tierarztliche Wochenschrift, 110(10), 426–428. [PubMed] [Google Scholar]
- Schwartz, K. J. (1991). Salmonellosis in swine. Compendium on Continuing Education for the Practising Veterinarian, 13, 139–146. [Google Scholar]
- Tizard, I. R. (2009). Immunity in the fetus and newborn In Tizard I. R. (Ed.), Veterinary Immunology and Immunopathology, 8th ed. (pp. 223–238). Philadelphia: Elsevier. [Google Scholar]
- Wills, R. W. , Gray, J. T. , Fedorka‐Cray, P. J. , Yoon, K. J. , Ladely, S. , & Zimmerman, J. J. (2000). Synergism between porcine reproductive and respiratory syndrome virus (PRRSV) and Salmonella choleraesuis in swine. Veterinary Microbiology, 71(3–4), 177–192. [DOI] [PMC free article] [PubMed] [Google Scholar]


