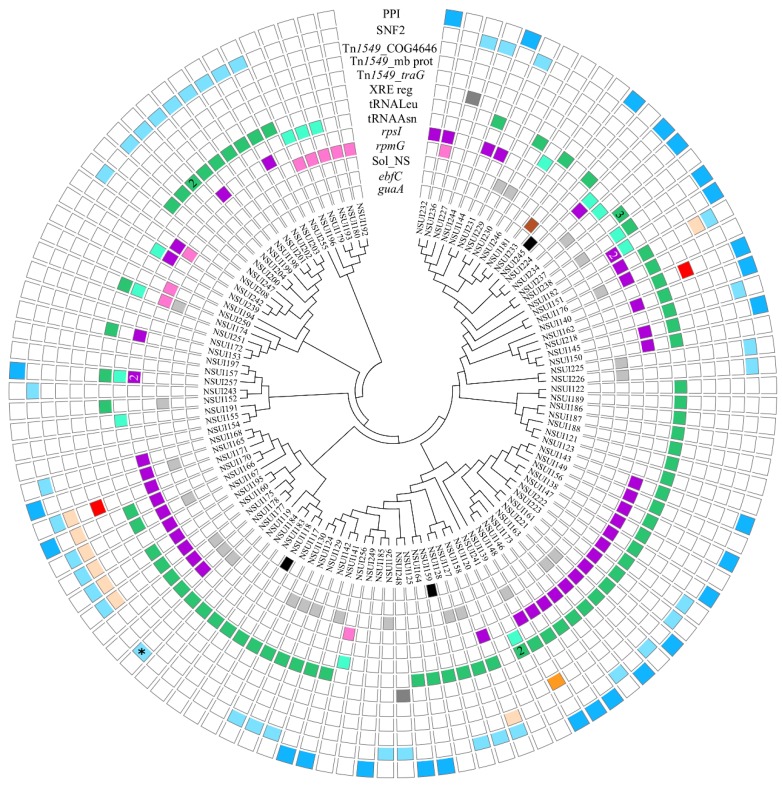
Figure 5.
Integrative and mobilizable elements detected in the genomes of S. suis strains of serotypes other than 2. Boxes located on the same line as a strain name are either empty (absence of IME) or colored (presence of an IME). Strains were grouped according to their MLST alleles (see material and method section for the method used to generate the cladogram that appears in the middle of the figure). The first two outer circles indicate IMEs integrated into the peptidylprolyl isomerase gene (in dark blue) and IMEs integrated into the SNF2 gene (in light blue) (IMEs integrated into ICEs belonging to the Tn5252 family). The third, 4th and 5th circles show IMEs integrated into ICEs of the Tn1549 family (from outside to inside: in a gene encoding a protein with a COG4646 domain shown with a salmon color, a membrane protein shown in orange and in traG shown in red). The next circle indicates the presence of IMEs integrated into an XRE regulator gene (dark grey color). Middle circles show IMEs integrated into the 3′ end of tRNA genes; in green for integration into tRNALeu gene (7th circle) and in light green for tRNAAsn gene (8th circle). On the following circles (still from outside to inside): IMEs integrated into rpsI (in purple on the 9th circle) and into rpmG (in pink on the 10th circle). Inner circles indicate IMEs with a low specificity of integration (in light grey on the 11th circle), in ebfC gene (in dark brown on the 12th circle) and in guaA (in black on the 13th last inner circle). One element resulting from recombination between an IME_SNF2 and an IME_PPI (called “hybrid” in Table S1) is indicated by a star. When several elements are present at the same integration site, the number of elements is indicated in the box (for example, 3 IME_tRNALeu in strain NSUI237).