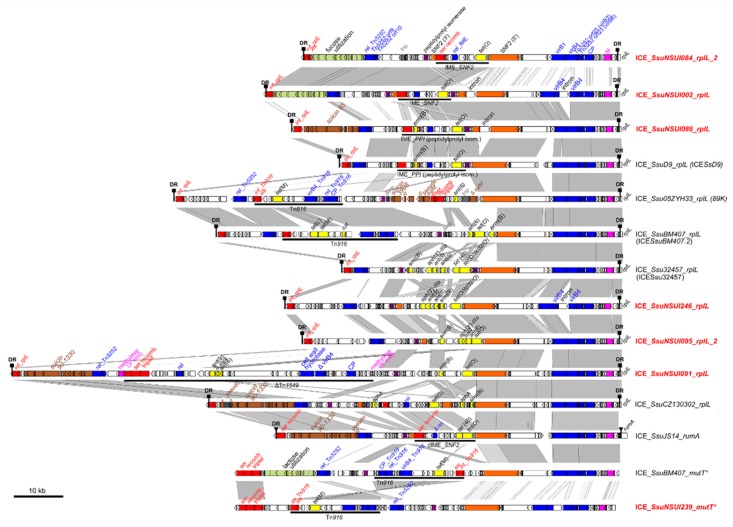
Figure 7.
Comparison of the integrative and conjugative elements (ICEs) belonging to the Tn5252 family and carrying AMR genes found in S. suis. ICEs are named according to their host strains and integration sites. When another name has already been published for the element, it is indicated in brackets. ICEs identified in this work are indicated in red. For more clarity, elements in accretion with ICEs of the Tn5252 family are not shown in the figure. ICEs or IMEs integrated inside ICEs are indicated as black lines with an indication of the name of the element. Nucleic acid sequence identity higher than 80% between two sequences is indicated in light gray and that higher than 90% in dark gray. Direct repeats (DR) delimiting ICEs are shown as squares or triangles depending on the integration gene (rplL, rumA or mutT *). Coding DNA sequences (CDSs) appear as arrows (truncated genes are indicated by deltas). Modules of recombination (integrase [int], excisionase [xis] and serine recombinase genes [ser recomb]) and conjugation appear in red and blue, respectively. The different integration sites (rplL, rumA, mutT *) targeted by the integrase are indicated in the figure and are part of the ICE name. Genes encoding proteins with putative function inferred from in silico analysis are indicated in yellow for antimicrobial resistance genes, in orange for SNF2, in purple for the peptidylprolyl isomerase gene, in green for sugar metabolism cluster, in pink for methylase genes, in gray for transposase genes, and in brown for bacteriocin synthesis clusters.