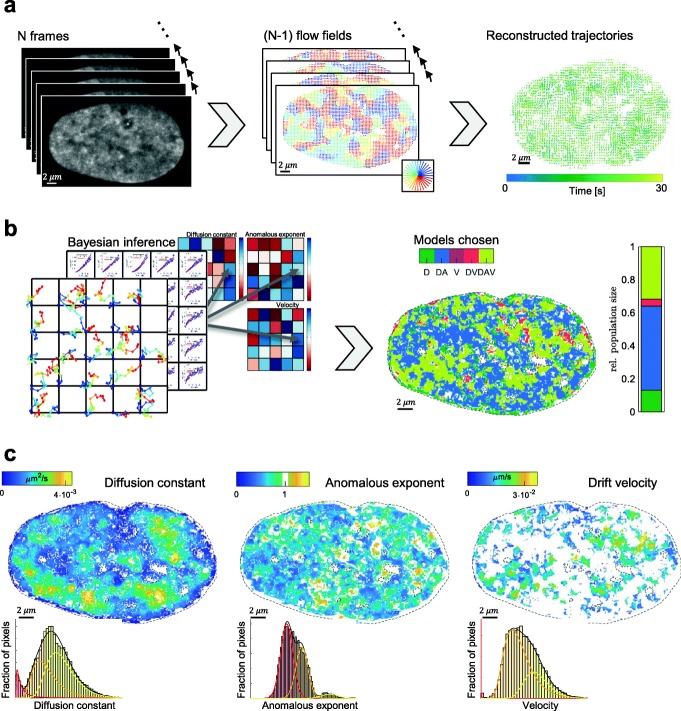
Fig. 1.
Hi-D enables spatially resolved mapping of genome dynamic properties at nanoscale resolution in living cells. Workflow: a A series of N = 150 confocal microscopy images acquired at 5 fps (left) (here SiR-DNA stained living U2OS cells). Dense optical flow was applied to define (N-1) flow fields of the images (center, color coded) based on fluorescence intensity of each pixel (size = 65 nm). Individual trajectories are reconstructed over the duration of acquisition (right). b MSD model selection (left): Trajectories of a 3 × 3 neighborhood of every pixel are used to calculate a mean MSD curve and its corresponding covariance matrix. By a Bayesian inference approach, the type of diffusion fitting each individual curve is chosen (free diffusion (D), anomalous diffusion (DA), directed motion (V), or a combination (DV) or (DAV). The spatial distribution of the selected models for each pixel is shown as a color map. c Maps of biophysical parameters (D, α, and V) extracted from the best describing model per pixel reveal local dynamic behavior of DNA in large domains. The distribution is deconvolved using a general mixture model