Figure 1.
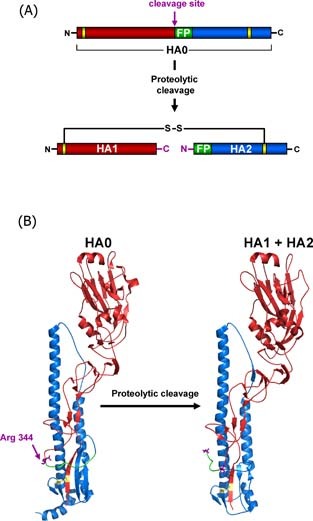
Structural rearrangements associated with influenza virus hemagglutinin cleavage by host cell proteases. The HA precursor HA0 is cleaved into two subunits HA1 (red) and HA2 (blue) by cellular proteases which recognise either a multibasic or monobasic cleavage site located in a loop between HA1 and HA2. Multibasic cleavage sites harbour multiple arginines and/or lysines and are found in the HA‐proteins of HPAIV. Monobasic cleavage sites consist of a single arginine (Arg 344 (purple) in case of the 1918 influenza virus) or lysine and are found in the HA‐proteins of human influenza viruses and LPAIV. Upon proteolytic cleavage, the fusion peptide (green) in HA2 is liberated from HA1 and the HA is primed for activation by low pH, which involves insertion of the fusion peptide into a negatively charged pocket located adjacent to the cleavage site‐bearing loop in HA0. The subunits HA1 and HA2 remain covalently linked by a single disulfide bond (yellow), the newly formed C‐terminus of HA1 and N‐terminus of HA2 are labelled in purple. (A) Schematic representation of HA cleavage. (B) Structural changes associated with cleavage of the 1918 influenza HA (A/South Carolina/1/18 (H1N1), Accession‐No: ADD17229, HA0 (1RD8.pdb), HA1 + HA2 (1RUZ.pdb)
