Figure 2.
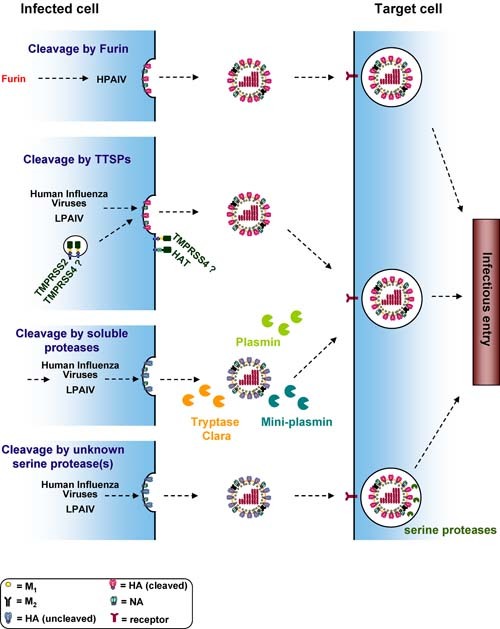
Activation of the influenza virus hemagglutinin in different cellular compartments. The influenza virus HA binds to alpha 2–3 linked (avian viruses) or alpha 2–6 linked (human viruses) sialic acids presented by proteins or lipids on the host cell surface. Upon endocytic uptake of virions, the acidic environment of the endosome triggers HA‐driven fusion of the viral and the endosomal membrane. After fusion the viral ribonucleoproteins are released into the host cell cytoplasm and transported to the nucleus, where viral genomic and messenger RNAs are synthesised. The viral membrane proteins HA, NA and M2 are synthesised in the secretory pathway. The HA proteins of HPAIV are cleaved in the Golgi apparatus/TGN by pro‐protein convertases of the subtilisin family, like furin or PC6, and cleaved HA is incorporated into progeny particles. The HA proteins of human influenza viruses and LPAIV can be cleaved by different proteases and in different cellular locations. HA can be transported to the cell surface, incorporated into virions in an uncleaved form and cleavage can subsequently be mediated by soluble proteases like plasmin in the extracellular space or by unknown serine protease(s) in endosomal vesicles of target cells. Alternatively, HA can be cleaved by the TTSPs TMPRSS2, TMPRSS4 and HAT, and cleavage could either occur en route to the cell membrane (TMPRSS2) or upon insertion into the target cell membrane (HAT). The cellular location where HA is cleaved by TMPRSS4 is at present unclear.
