Figure 1. A Microbial Factory: Generation of Genome-wide crRNA Libraries by CRISPR-Cas Adaptation.
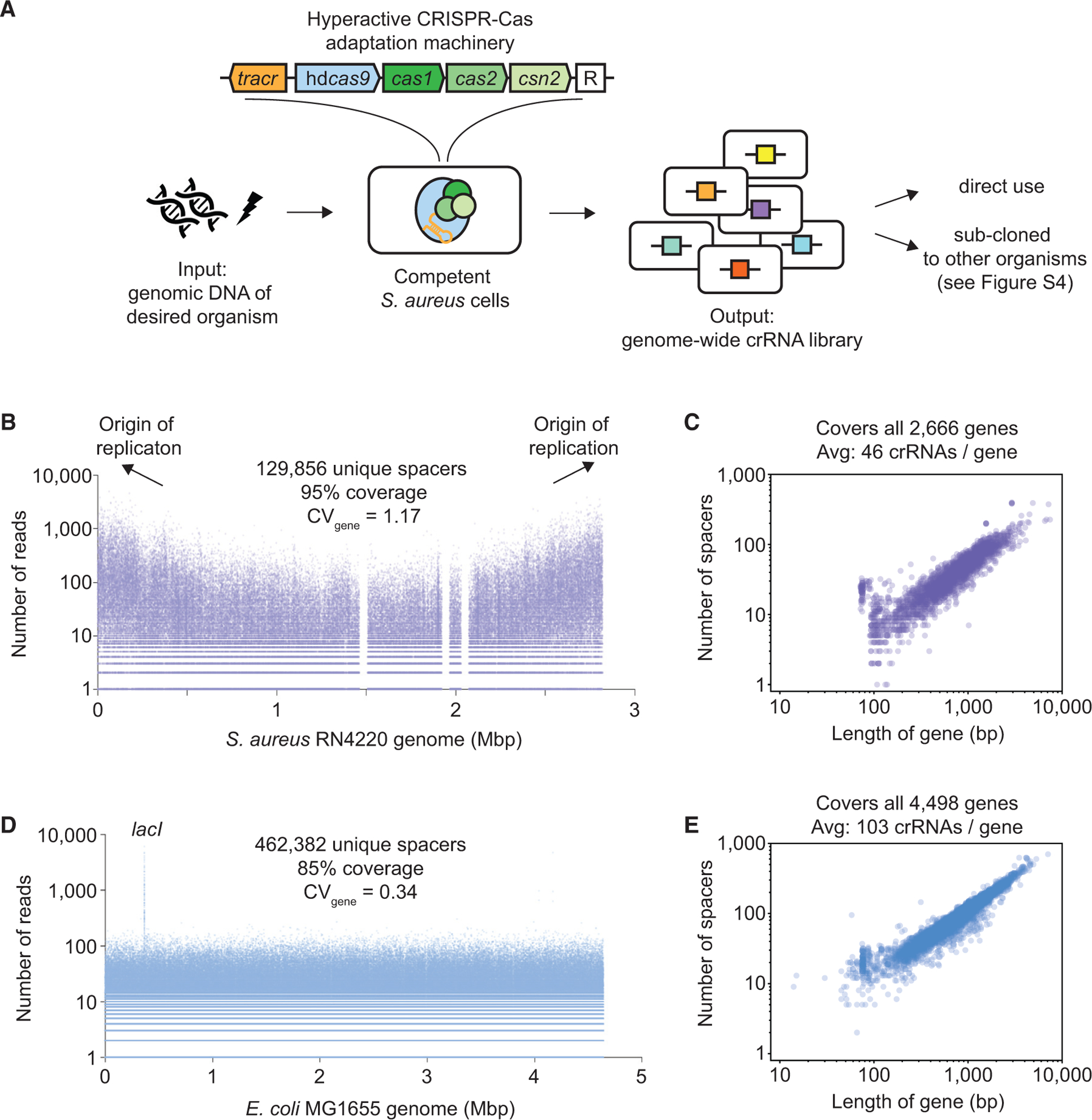
(A) CRISPR adaptation-mediated library manufacturing (CALM). A hyperactive CRISPR-Cas adaptation machinery consists of the 89-nt tracrRNA, hdCas9, Cas1, Cas2, and Csn2. New spacers are integrated into the empty CRISPR array, denoted as “R.” To generate a diverse crRNA library, sheared genomic DNA is electroporated into competent S. aureus cells harboring the adaptation machinery.
(B) A crRNA library was generated by electroporating S. aureus RN4220 genomic DNA as described in (A). The number of reads and location of all 129,856 sequenced spacers matching the genome are shown. The genome contains 136,928 PAMs. Three gap regions correspond to prophages present in the NCBI reference genome (NCTC8325) but missing in RN4220.
(C) Number of spacers mapped to each of all 2,666 annotated genes in S. aureus RN4220 versus gene length.
(D) A crRNA library was generated by electroporating E. coli MG1655 genomic DNA as described in Figure S4A. The number of reads and location of all 462,382 sequenced spacers matching the genome are shown. The genome contains 542,073 PAMs. lacI was preferentially enriched because of an additional presence of the gene in a helper plasmid, pCCC (Figure S4A).
(E) Number of spacers mapped to each of all 4,498 annotated genes in E. coli MG1655 versus gene length.
