Figure 2.
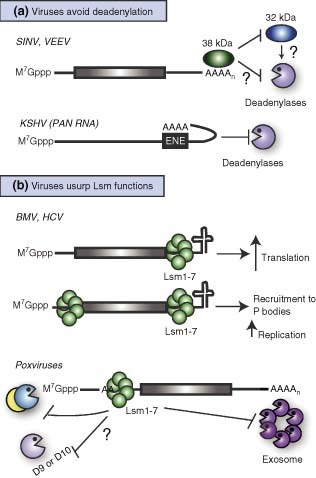
Viral circumvention or utilization of cellular deadenylation and decapping pathways. (a) The RNAs of Sindbis virus (SINV) and Venezuelan equine encephalitis virus (VEEV) contains elements in the 3′ UTR that recruit a cellular 38‐kDa protein and block deadenylation, presumably by preventing binding of a 32‐kDa deadenylation‐promoting factor. The PAN RNA of Kaposi's sarcoma‐associated herpesvirus contains an element (ENE) that interacts in cis with the poly(A) tail thus preventing deadenylase access. (b) The RNA genomes of Brome mosaic virus (BMV) and hepatitis C virus (HCV) can be bound by Lsm proteins in their 3′ and/or 5′ UTRs. This enhances their translation, as well as facilitates replication, possibly through recruitment of the RNAs to P bodies. Poxviruses transcripts have polyadenine sequences within their 5′ UTRs, which bind Lsm1–7. Lsm binding prevents decapping (and presumably 5′→3′ decay), as well as 3′→5′ degradation of the messages.
