Figure 3.
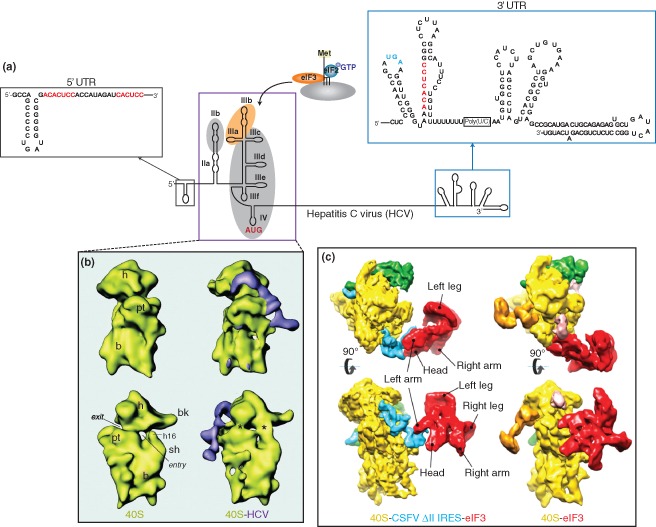
Hepatitis C virus (HCV) IRES. (a) Genome organization of the hepatitis C virus. The 5′‐untranslated region of the HCV genome contains an internal ribosome entry site (boxed in purple) that mediates ribosome recruitment using only eIF3 and the eIF2·Met‐tRNAi·GTP ternary complex. The IRES consists of domains II–IV, where the apical region of domain III interacts with eIF3 (shaded in orange), and regions within domains II, III, and IV establish contacts with the ribosome (shaded in gray). The IRES translational start site is highlighted in red. Within the 5′ and 3′ untranslated regions (UTRs) of the viral genome, three miR‐122 binding sites have been identified by base complementarity (sequences highlighted in red). The stop codon of the coding region is highlighted in blue. The structures of the HCV 5′ and 3′ UTRs are Reprinted with permission from Ref 64. Copyright 2005 American Association for the Advancement of Science. (b) Cryo‐EM structure of the vacant 40S ribosomal subunit from rabbit reticulocytes and 40S‐HCV complex at 20Å. The HCV IRES binds to the solvent accessible side of the ribosome and induces conformational changes in the 40S subunit, similar to those induced by IGR IRES‐40S binding (indicated by asterisks). (Reprinted with permission from Ref 65. Copyright 2001 American Association for the Advancement of Science). (c) Cryo‐EM structure of the 40S‐eIF3 (11.6 Å) and 40S‐CSFV ΔII IRES‐eIF3 (9.5 Å) complexes containing eIF3 and 40S from rabbit reticulocytes. In the 40S‐eIF3 complex (right), eIF3 (shown in red) interacts directly with the 40S subunit (in yellow). In contrast, eIF3 binds to the IRES (in blue) via the apical loop of domain III in the IRES‐containing complex (left), suggesting that the IRES displaces eIF3 to gain access to the ribosome. (Reprinted with permission from Ref 66. Copyright 2013 Nature Publishing Group)
