Figure 3.
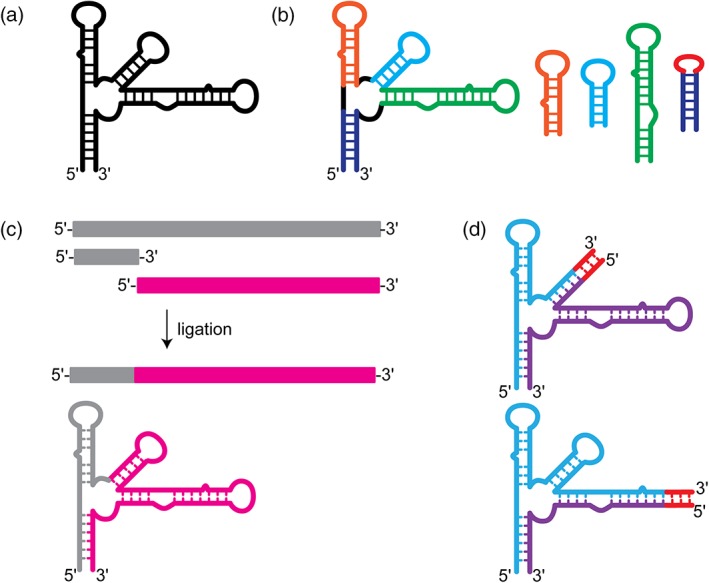
Methods for fragmentation or segmental labeling of large RNAs. (a) Secondary structure of a large RNA with a complex multi‐helix junction. (b) “Divide and conquer” approach. Small oligo RNAs are designed to mimic the structure of particular regions within the large RNA. These oligo RNAs are well‐suited for rapid chemical shift assignment by traditional methods. (c) Enzymatic ligation allows for segmental labeling of a large RNAs. Two RNAs are independently synthesized and can therefore be independently labeled. The two RNAs can then be ligated together to make a full‐length construct with only one region of the RNA containing nuclear magnetic resonance‐active isotope labeling. (d) Fragmentation‐based segmental labeling. The RNA of interest is fragmented at a hairpin loop. The loop sequence is replaced (in some cases) with a run of intermolecular C–G base pairs which serve as an annealing handle. This approach in theory can be applied at any hairpin loop structure within a large RNA molecules
